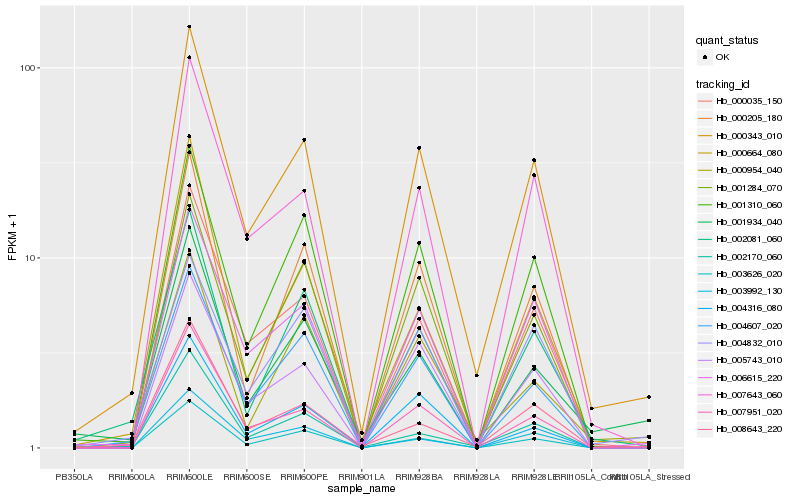
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_010 |
0.0 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 2 |
Hb_006615_220 |
0.0895665864 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000035_150 |
0.0990885115 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 4 |
Hb_007643_060 |
0.0991219615 |
- |
- |
- |
| 5 |
Hb_007951_020 |
0.1071909702 |
- |
- |
cationic amino acid transporter 5 family protein [Populus trichocarpa] |
| 6 |
Hb_004607_020 |
0.1090099345 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 7 |
Hb_003626_020 |
0.1094703039 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 8 |
Hb_001310_060 |
0.1111175153 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 9 |
Hb_005743_010 |
0.1112840263 |
- |
- |
RING/U-box superfamily protein, putative [Theobroma cacao] |
| 10 |
Hb_000205_180 |
0.1119907201 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 11 |
Hb_008643_220 |
0.1135798988 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 12 |
Hb_002081_060 |
0.1167767759 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 13 |
Hb_001284_070 |
0.1216641773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000664_080 |
0.1257378364 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X2 [Jatropha curcas] |
| 15 |
Hb_002170_060 |
0.1266101928 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_004316_080 |
0.1272089745 |
- |
- |
PREDICTED: uncharacterized protein LOC104602212 [Nelumbo nucifera] |
| 17 |
Hb_003992_130 |
0.1301062498 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000954_040 |
0.1324734268 |
- |
- |
PREDICTED: metal tolerance protein 4 [Jatropha curcas] |
| 19 |
Hb_001934_040 |
0.1329046414 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 20 |
Hb_004832_010 |
0.1342376293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |