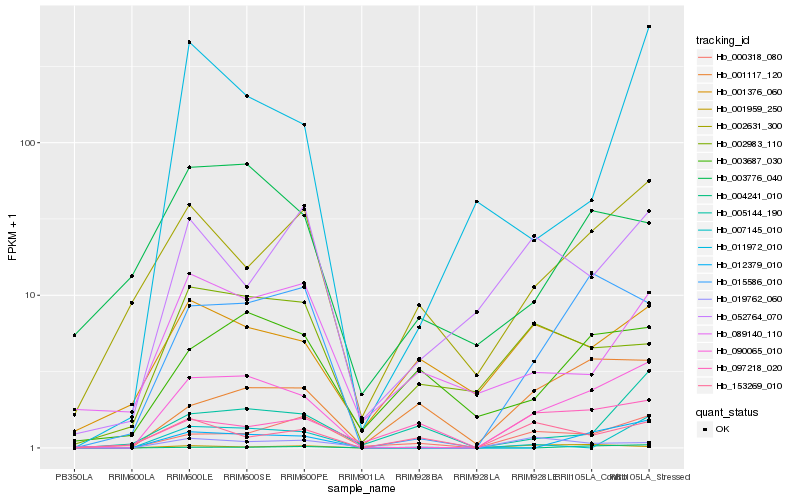
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090065_010 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_015586_010 |
0.1992590744 |
- |
- |
PREDICTED: flavonol sulfotransferase-like [Jatropha curcas] |
| 3 |
Hb_012379_010 |
0.2090404134 |
- |
- |
- |
| 4 |
Hb_000318_080 |
0.2696420043 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 5 |
Hb_002631_300 |
0.2709261511 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 6 |
Hb_003687_030 |
0.2778988851 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 7 |
Hb_001117_120 |
0.2859288912 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 8 |
Hb_011972_010 |
0.2877324982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007145_010 |
0.2935589999 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 10 |
Hb_003776_040 |
0.297455063 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 11 |
Hb_005144_190 |
0.2978814391 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 12 |
Hb_004241_010 |
0.3010330085 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_052764_070 |
0.3017329849 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 14 |
Hb_001376_060 |
0.3021850096 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |
| 15 |
Hb_019762_060 |
0.3024913097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002983_110 |
0.3025442406 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 17 |
Hb_089140_110 |
0.3083281129 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_153269_010 |
0.3083792557 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 19 |
Hb_001959_250 |
0.3123382651 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 20 |
Hb_097218_020 |
0.3130273717 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |