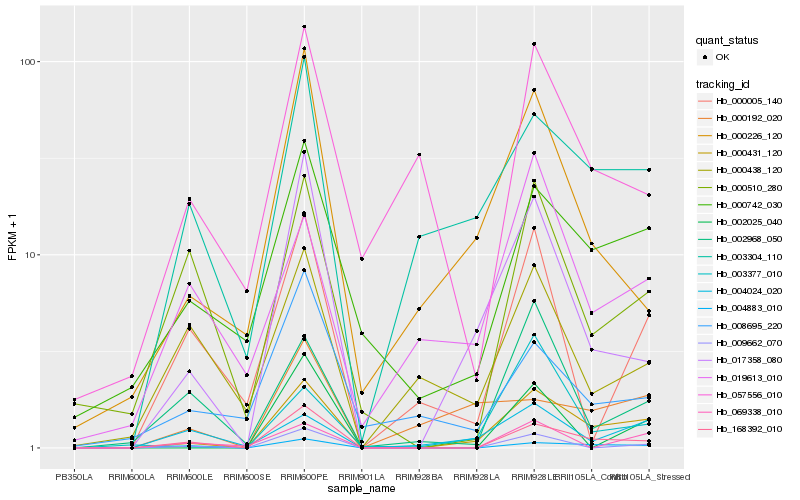
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000431_120 |
0.0 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_168392_010 |
0.1901347261 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_004883_010 |
0.2088551024 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 4 |
Hb_017358_080 |
0.2123661813 |
- |
- |
- |
| 5 |
Hb_003304_110 |
0.2297358872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000742_030 |
0.232726854 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 7 |
Hb_000226_120 |
0.2618069537 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009662_070 |
0.2665577623 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 9 |
Hb_004024_020 |
0.2720404196 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003377_010 |
0.272072304 |
- |
- |
hypothetical protein POPTR_0016s06140g, partial [Populus trichocarpa] |
| 11 |
Hb_000510_280 |
0.2765818545 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 12 |
Hb_002968_050 |
0.2769556447 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 13 |
Hb_002025_040 |
0.2825293343 |
- |
- |
PREDICTED: WW domain-binding protein 4 [Jatropha curcas] |
| 14 |
Hb_000005_140 |
0.2866617471 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 15 |
Hb_000192_020 |
0.2890769421 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 16 |
Hb_019613_010 |
0.2942883828 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_069338_010 |
0.2959176954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008695_220 |
0.2968963936 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 19 |
Hb_057556_010 |
0.2996138856 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 20 |
Hb_000438_120 |
0.3007081245 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |