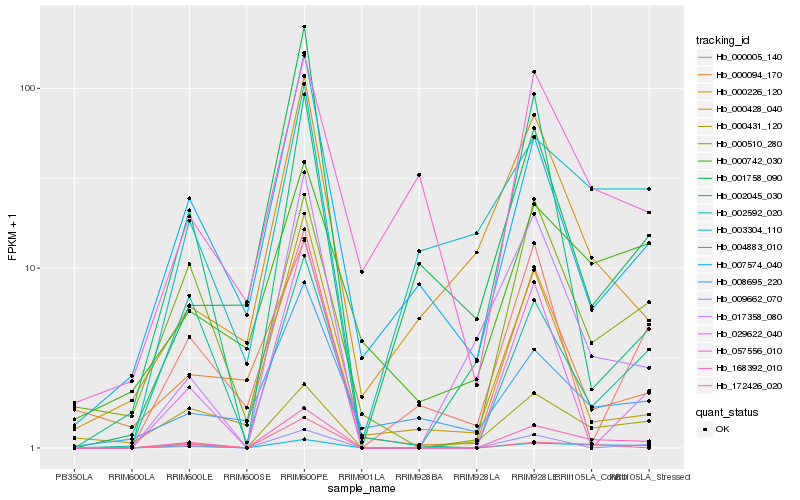
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168392_010 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_004883_010 |
0.1315696115 |
- |
- |
hypothetical protein JCGZ_23144 [Jatropha curcas] |
| 3 |
Hb_000431_120 |
0.1901347261 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_017358_080 |
0.2129661844 |
- |
- |
- |
| 5 |
Hb_029622_040 |
0.2208261329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000510_280 |
0.2227524081 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 7 |
Hb_009662_070 |
0.2266378377 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 8 |
Hb_007574_040 |
0.2437448799 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 9 |
Hb_000742_030 |
0.251212206 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 10 |
Hb_000428_040 |
0.2526080398 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 11 |
Hb_003304_110 |
0.2561223501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002592_020 |
0.2571718967 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008695_220 |
0.2581911113 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 14 |
Hb_000094_170 |
0.2588495286 |
- |
- |
PREDICTED: uncharacterized protein LOC105633781 [Jatropha curcas] |
| 15 |
Hb_000226_120 |
0.261182398 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_172426_020 |
0.2637577207 |
- |
- |
PREDICTED: putative fasciclin-like arabinogalactan protein 20 [Jatropha curcas] |
| 17 |
Hb_002045_030 |
0.2679884304 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein CISIN_1g047687mg [Citrus sinensis] |
| 18 |
Hb_001758_090 |
0.2711796493 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 19 |
Hb_057556_010 |
0.2731385915 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 20 |
Hb_000005_140 |
0.273915659 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |