| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
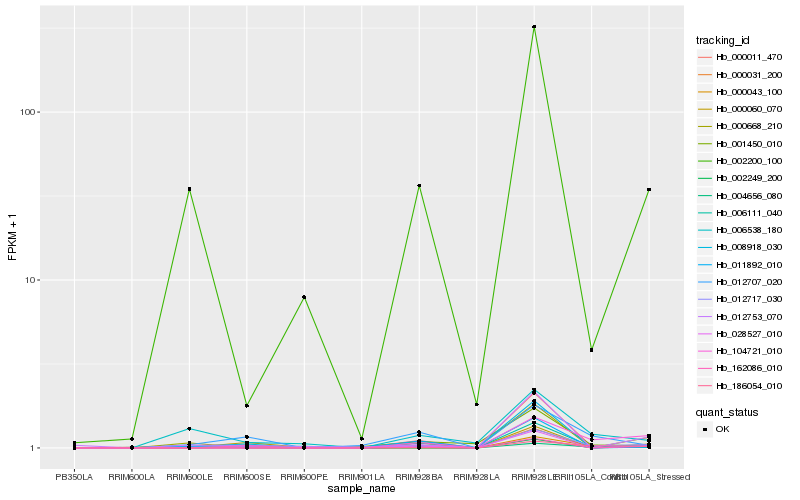
Hb_012717_030 |
0.0 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 2 |
Hb_104721_010 |
0.2372187678 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 3 |
Hb_000011_470 |
0.2413033461 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 4 |
Hb_006111_040 |
0.2442738399 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 5 |
Hb_000060_070 |
0.2448399551 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 6 |
Hb_012707_020 |
0.2457757928 |
- |
- |
- |
| 7 |
Hb_002200_100 |
0.2550094637 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 8 |
Hb_000043_100 |
0.255680832 |
- |
- |
PREDICTED: uncharacterized protein LOC105797583 [Gossypium raimondii] |
| 9 |
Hb_162086_010 |
0.2644002937 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 10 |
Hb_186054_010 |
0.264735098 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_000668_210 |
0.2652003817 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 12 |
Hb_011892_010 |
0.2695622593 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_012753_070 |
0.2695647566 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 14 |
Hb_008918_030 |
0.2733077036 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 15 |
Hb_000031_200 |
0.2740277486 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM3-like [Jatropha curcas] |
| 16 |
Hb_028527_010 |
0.2742387835 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 17 |
Hb_006538_180 |
0.2753827718 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger family protein [Populus trichocarpa] |
| 18 |
Hb_004656_080 |
0.2758367393 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_001450_010 |
0.2762791991 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 20 |
Hb_002249_200 |
0.2798892675 |
transcription factor |
TF Family: LFY |
LEAFY-like protein [Pistacia chinensis] |