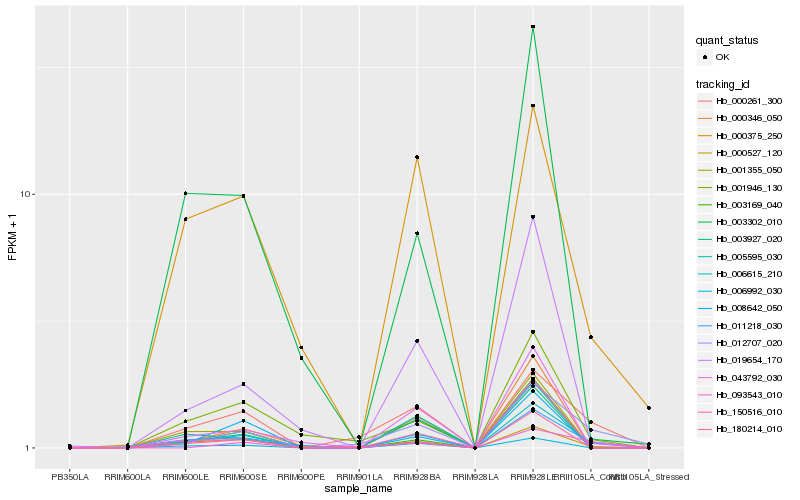
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006615_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640677 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012707_020 |
0.1919885192 |
- |
- |
- |
| 3 |
Hb_000261_300 |
0.1934489503 |
- |
- |
- |
| 4 |
Hb_001355_050 |
0.1934595527 |
- |
- |
- |
| 5 |
Hb_003927_020 |
0.198541269 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 6 |
Hb_011218_030 |
0.205031687 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |
| 7 |
Hb_001946_130 |
0.207631805 |
- |
- |
- |
| 8 |
Hb_019654_170 |
0.2082148985 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 9 |
Hb_006992_030 |
0.2112885512 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000346_050 |
0.2141010045 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000527_120 |
0.2189032869 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 12 |
Hb_150516_010 |
0.2199104074 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_008642_050 |
0.2216891684 |
- |
- |
- |
| 14 |
Hb_093543_010 |
0.2243352364 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 15 |
Hb_005595_030 |
0.2254712862 |
- |
- |
hypothetical protein EUGRSUZ_A00624 [Eucalyptus grandis] |
| 16 |
Hb_000375_250 |
0.2255162761 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 17 |
Hb_003169_040 |
0.2269795986 |
- |
- |
- |
| 18 |
Hb_180214_010 |
0.2273622261 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 19 |
Hb_043792_030 |
0.2280616601 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 20 |
Hb_003302_010 |
0.2301004849 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |