| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
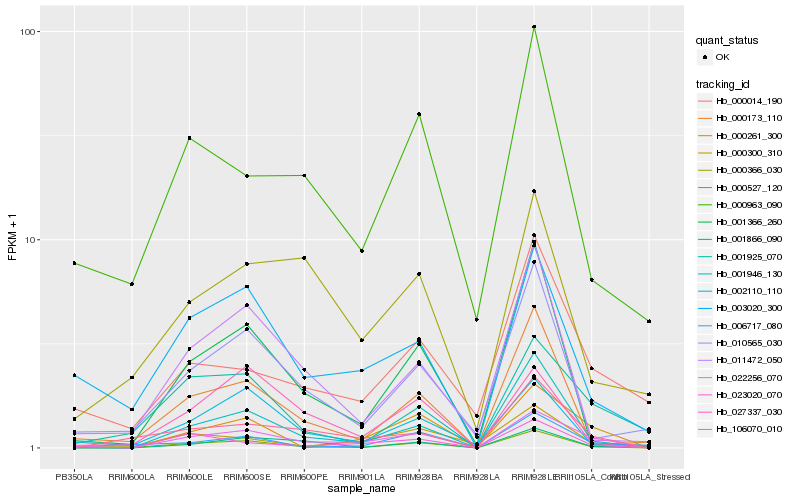
Hb_022256_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 2 |
Hb_000527_120 |
0.1865325444 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 3 |
Hb_000261_300 |
0.1940559879 |
- |
- |
- |
| 4 |
Hb_023020_070 |
0.1998588538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003020_300 |
0.2073492522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001946_130 |
0.2098042773 |
- |
- |
- |
| 7 |
Hb_106070_010 |
0.2229751915 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 8 |
Hb_001866_090 |
0.2294897539 |
- |
- |
hypothetical protein VITISV_023491 [Vitis vinifera] |
| 9 |
Hb_000014_190 |
0.2369989965 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_001925_070 |
0.2378290557 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Populus euphratica] |
| 11 |
Hb_002110_110 |
0.2387099835 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 12 |
Hb_001366_260 |
0.2414496615 |
- |
- |
T4.15 [Malus x robusta] |
| 13 |
Hb_000963_090 |
0.2416299823 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 14 |
Hb_000300_310 |
0.2417756029 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_000173_110 |
0.2422698972 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 16 |
Hb_027337_030 |
0.2436238426 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 17 |
Hb_006717_080 |
0.2438122243 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 18 |
Hb_011472_050 |
0.2467102953 |
- |
- |
- |
| 19 |
Hb_010565_030 |
0.2481704684 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 20 |
Hb_000366_030 |
0.2486747513 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |