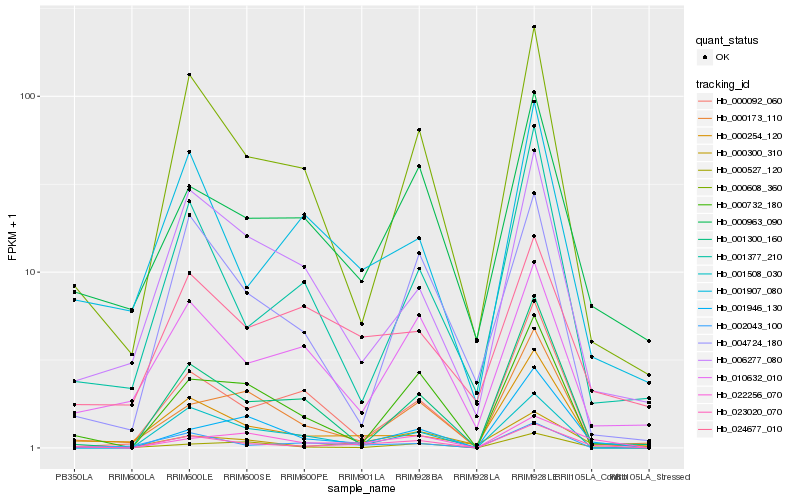
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023020_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_022256_070 |
0.1998588538 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 3 |
Hb_000300_310 |
0.204396388 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000608_360 |
0.2044231586 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 5 |
Hb_000254_120 |
0.2130523477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000963_090 |
0.2205767271 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_000527_120 |
0.2217750283 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_001907_080 |
0.2249857469 |
- |
- |
PREDICTED: uncharacterized protein LOC105628749 [Jatropha curcas] |
| 9 |
Hb_000732_180 |
0.233895565 |
- |
- |
Uncharacterized protein TCM_030007 [Theobroma cacao] |
| 10 |
Hb_002043_100 |
0.2341353315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001508_030 |
0.2370330173 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 12 |
Hb_006277_080 |
0.2381511457 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 13 |
Hb_001300_160 |
0.2382216412 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 14 |
Hb_001946_130 |
0.2427409573 |
- |
- |
- |
| 15 |
Hb_004724_180 |
0.2433160421 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 16 |
Hb_000173_110 |
0.2436874232 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 17 |
Hb_010632_010 |
0.2440550651 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001377_210 |
0.2442528642 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_024677_010 |
0.2454180438 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 20 |
Hb_000092_060 |
0.2454452887 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |