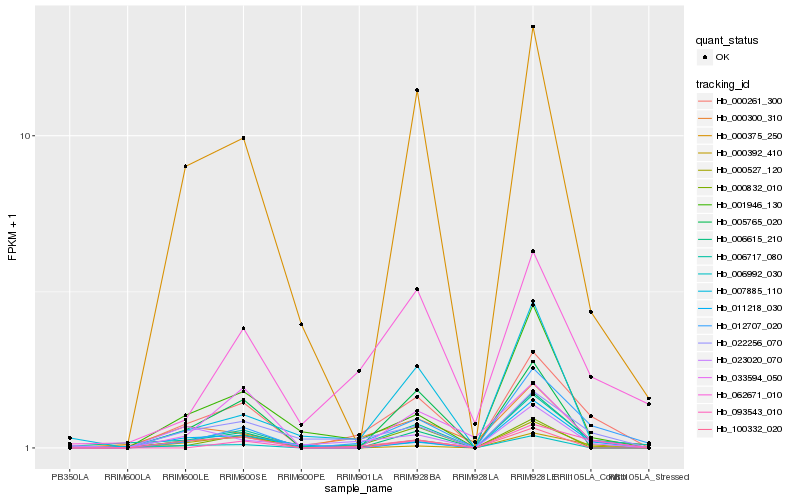
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_012707_020 |
0.1768339102 |
- |
- |
- |
| 3 |
Hb_006615_210 |
0.1934489503 |
- |
- |
PREDICTED: uncharacterized protein LOC105640677 isoform X2 [Jatropha curcas] |
| 4 |
Hb_022256_070 |
0.1940559879 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 5 |
Hb_006717_080 |
0.2268322496 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 6 |
Hb_000527_120 |
0.2398655094 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 7 |
Hb_000375_250 |
0.2485495906 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |
| 8 |
Hb_000300_310 |
0.2550403711 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_001946_130 |
0.2558693243 |
- |
- |
- |
| 10 |
Hb_100332_020 |
0.2564749494 |
- |
- |
PREDICTED: uncharacterized protein LOC105168695 [Sesamum indicum] |
| 11 |
Hb_062671_010 |
0.2584160661 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_093543_010 |
0.2650617291 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_023020_070 |
0.2750309337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007885_110 |
0.2790916395 |
- |
- |
PREDICTED: uncharacterized protein LOC104109376 [Nicotiana tomentosiformis] |
| 15 |
Hb_000392_410 |
0.2807970351 |
- |
- |
PREDICTED: uncharacterized protein LOC104882159 [Vitis vinifera] |
| 16 |
Hb_000832_010 |
0.2866603428 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 17 |
Hb_005765_020 |
0.2877164636 |
- |
- |
- |
| 18 |
Hb_006992_030 |
0.288598443 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_033594_050 |
0.2886992528 |
- |
- |
PREDICTED: cyclin-D4-1-like [Jatropha curcas] |
| 20 |
Hb_011218_030 |
0.2902306034 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |