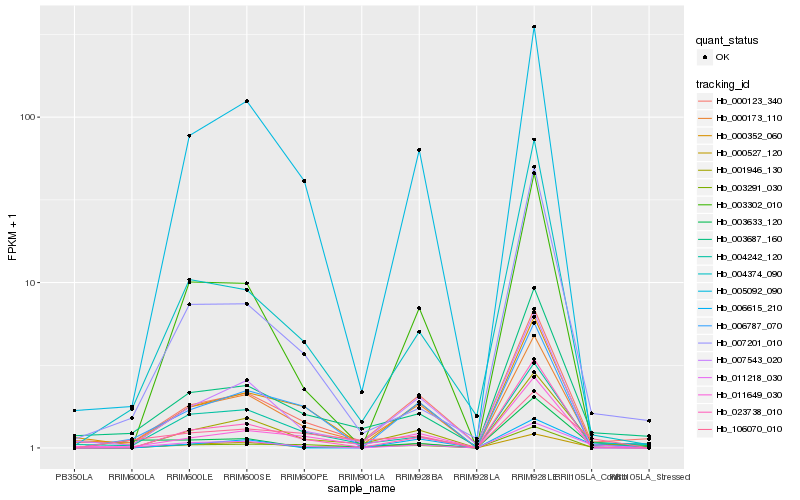
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_030 |
0.0 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |
| 2 |
Hb_001946_130 |
0.1654747447 |
- |
- |
- |
| 3 |
Hb_000527_120 |
0.1856594014 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 4 |
Hb_003302_010 |
0.2002678477 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 5 |
Hb_003687_160 |
0.2012239228 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_003291_030 |
0.2027841391 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 7 |
Hb_006615_210 |
0.205031687 |
- |
- |
PREDICTED: uncharacterized protein LOC105640677 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000123_340 |
0.2109772555 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 9 |
Hb_004374_090 |
0.2116415294 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 10 |
Hb_106070_010 |
0.2122016986 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 11 |
Hb_007201_010 |
0.2161844103 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004242_120 |
0.2175940013 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 13 |
Hb_023738_010 |
0.2179931673 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 14 |
Hb_000352_060 |
0.2190139598 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 15 |
Hb_003633_120 |
0.2204114261 |
- |
- |
PREDICTED: uncharacterized protein LOC104880778 [Vitis vinifera] |
| 16 |
Hb_000173_110 |
0.2207001921 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 17 |
Hb_005092_090 |
0.22109434 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 18 |
Hb_006787_070 |
0.224242202 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 19 |
Hb_007543_020 |
0.2243539182 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 20 |
Hb_011649_030 |
0.2246764119 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |