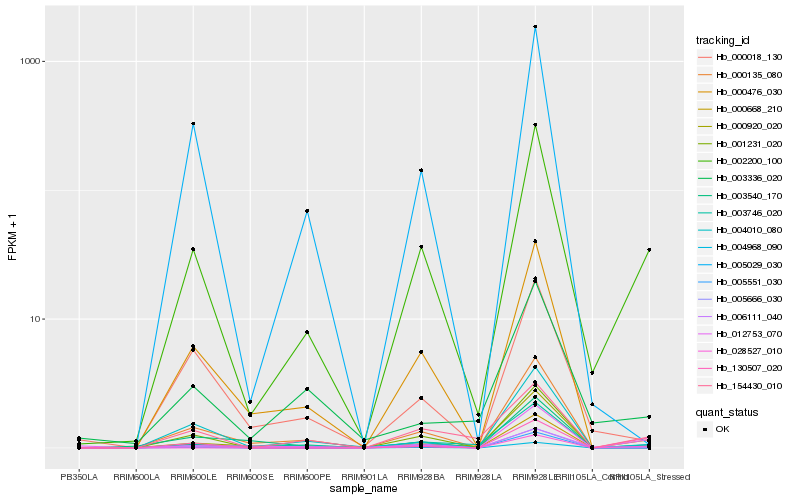
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002200_100 |
0.0 |
- |
- |
PREDICTED: pathogen-related protein-like [Jatropha curcas] |
| 2 |
Hb_000668_210 |
0.1474168387 |
- |
- |
hypothetical protein PHAVU_006G031500g [Phaseolus vulgaris] |
| 3 |
Hb_154430_010 |
0.1928016108 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003746_020 |
0.2002252805 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Gossypium raimondii] |
| 5 |
Hb_004010_080 |
0.2008654781 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_006111_040 |
0.2136965488 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 7 |
Hb_130507_020 |
0.2157795241 |
- |
- |
- |
| 8 |
Hb_012753_070 |
0.2171088998 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 9 |
Hb_000920_020 |
0.2227155375 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 10 |
Hb_000476_030 |
0.2230024886 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 11 |
Hb_005029_030 |
0.2254470104 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 12 |
Hb_003540_170 |
0.2264869866 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000135_080 |
0.2265501857 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_000018_130 |
0.2309602942 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 15 |
Hb_005666_030 |
0.2323340969 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 16 |
Hb_005551_030 |
0.2324175944 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 17 |
Hb_004968_090 |
0.2326724291 |
- |
- |
PREDICTED: uncharacterized protein LOC104882781 [Vitis vinifera] |
| 18 |
Hb_001231_020 |
0.2334032036 |
- |
- |
- |
| 19 |
Hb_028527_010 |
0.2340158788 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 20 |
Hb_003336_020 |
0.2340819183 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |