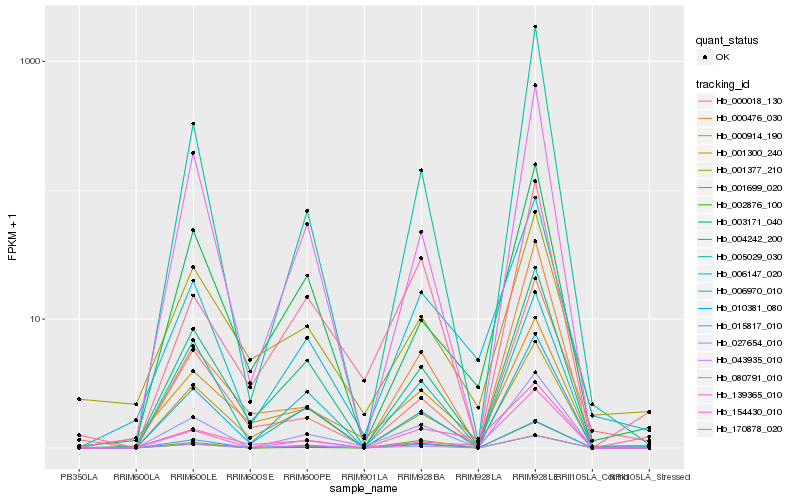
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006970_010 |
0.0 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 2 |
Hb_000914_190 |
0.1551782573 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_139365_010 |
0.159131626 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 4 |
Hb_027654_010 |
0.1613680774 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 5 |
Hb_015817_010 |
0.1693868027 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 6 |
Hb_004242_200 |
0.1705522286 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 7 |
Hb_001699_020 |
0.1708768837 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 8 |
Hb_001377_210 |
0.1774201948 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 9 |
Hb_010381_080 |
0.1791588811 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_003171_040 |
0.1806983166 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_006147_020 |
0.1818115252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_043935_010 |
0.1818743974 |
- |
- |
Receptor protein kinase-like protein [Medicago truncatula] |
| 13 |
Hb_005029_030 |
0.1822020709 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 14 |
Hb_000476_030 |
0.1885948631 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 15 |
Hb_080791_010 |
0.1889982021 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 16 |
Hb_000018_130 |
0.1899528059 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 17 |
Hb_001300_240 |
0.1905495071 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_154430_010 |
0.1914544534 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X2 [Jatropha curcas] |
| 19 |
Hb_170878_020 |
0.1932213315 |
- |
- |
- |
| 20 |
Hb_002876_100 |
0.1952619658 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |