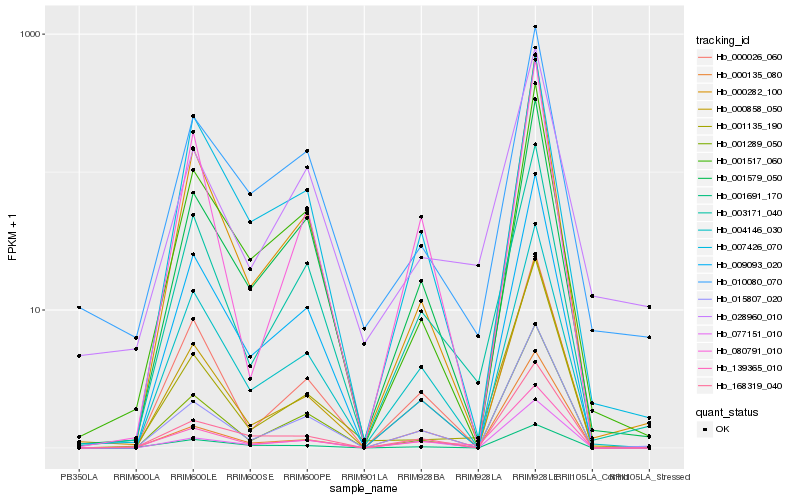
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139365_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 2 |
Hb_003171_040 |
0.1105170402 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_168319_040 |
0.1151768056 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 4 |
Hb_000858_050 |
0.1188349121 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_015807_020 |
0.1249710039 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_001579_050 |
0.1270666569 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_004146_030 |
0.1284573734 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000026_060 |
0.1357291647 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |
| 9 |
Hb_077151_010 |
0.1385512254 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 10 |
Hb_000282_100 |
0.142411854 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 11 |
Hb_001289_050 |
0.1444970233 |
- |
- |
hypothetical protein Poptr_cp075 [Populus trichocarpa] |
| 12 |
Hb_001135_190 |
0.1446629936 |
transcription factor |
TF Family: Orphans |
Pseudo response regulator isoform 11 [Theobroma cacao] |
| 13 |
Hb_001517_060 |
0.1448091175 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 14 |
Hb_010080_070 |
0.1454097207 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001691_170 |
0.1457456698 |
- |
- |
PREDICTED: callose synthase 12-like [Glycine max] |
| 16 |
Hb_028960_010 |
0.1463657935 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 17 |
Hb_007426_070 |
0.1471750857 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 18 |
Hb_009093_020 |
0.1490434229 |
- |
- |
PREDICTED: uncharacterized protein LOC105642100 [Jatropha curcas] |
| 19 |
Hb_080791_010 |
0.1493127638 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 20 |
Hb_000135_080 |
0.1507699384 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |