| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
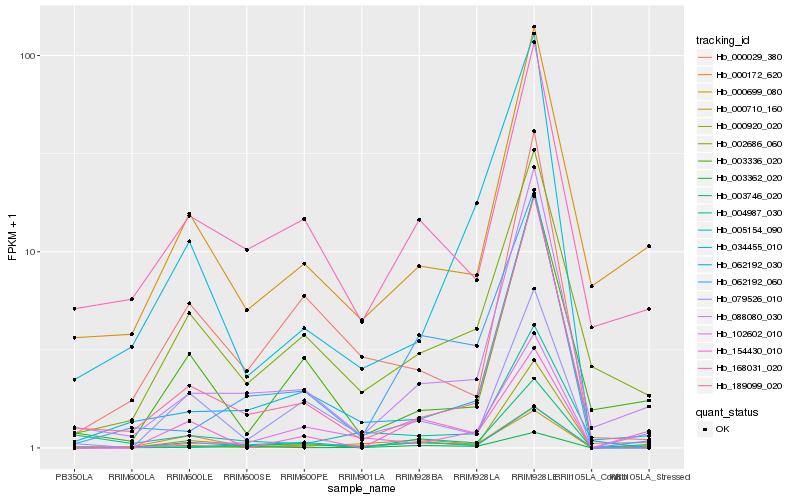
Hb_000710_160 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_003746_020 |
0.1508635048 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Gossypium raimondii] |
| 3 |
Hb_062192_060 |
0.1860739796 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 4 |
Hb_088080_030 |
0.1866153685 |
- |
- |
ribosomal protein S4 [Hevea brasiliensis] |
| 5 |
Hb_002686_060 |
0.1877075747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000699_080 |
0.1934156512 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000920_020 |
0.2026742723 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 8 |
Hb_000172_620 |
0.2036647621 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 9 |
Hb_079526_010 |
0.2050796515 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Populus euphratica] |
| 10 |
Hb_003362_020 |
0.2105575881 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 11 |
Hb_168031_020 |
0.2133793699 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 12 |
Hb_062192_030 |
0.2136311927 |
- |
- |
ribosomal protein S1 (mitochondrion) [Hevea brasiliensis] |
| 13 |
Hb_154430_010 |
0.215412981 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X2 [Jatropha curcas] |
| 14 |
Hb_005154_090 |
0.2164680529 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_004987_030 |
0.2184644218 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 16 |
Hb_189099_020 |
0.2187107488 |
- |
- |
NADH dehydrogenase subunit 9 (mitochondrion) [Hevea brasiliensis] |
| 17 |
Hb_000029_380 |
0.2200334588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003336_020 |
0.2226873359 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 19 |
Hb_102602_010 |
0.223815864 |
- |
- |
orf175 (mitochondrion) [Panax ginseng] |
| 20 |
Hb_034455_010 |
0.2248978315 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |