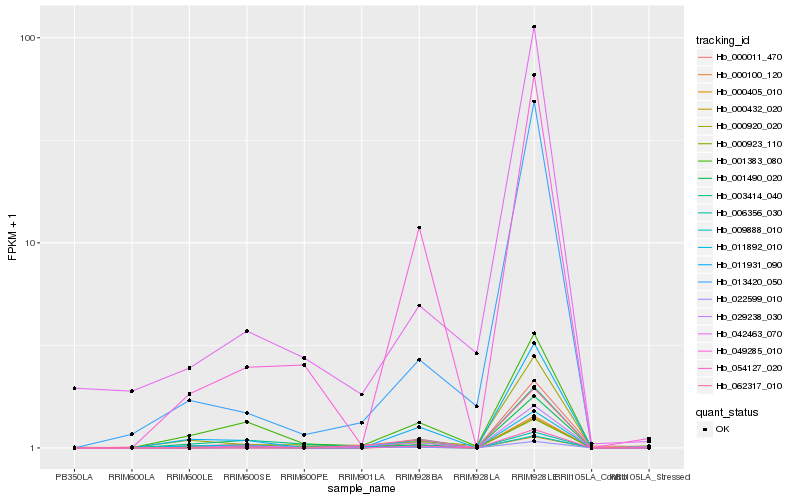
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029238_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103489421 [Cucumis melo] |
| 2 |
Hb_001490_020 |
0.1485842815 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_000011_470 |
0.1536012474 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 4 |
Hb_011892_010 |
0.1566645942 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_003414_040 |
0.1621271172 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 6 |
Hb_049285_010 |
0.171036971 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 7 |
Hb_000405_010 |
0.1795458737 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 8 |
Hb_000920_020 |
0.1831552061 |
- |
- |
OSJNBa0004L19.22 [Oryza sativa Japonica Group] |
| 9 |
Hb_000432_020 |
0.1840188321 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_054127_020 |
0.189685056 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 11 |
Hb_062317_010 |
0.1897128874 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_022599_010 |
0.1933313278 |
- |
- |
Uncharacterized protein TCM_010507 [Theobroma cacao] |
| 13 |
Hb_011931_090 |
0.1933554026 |
- |
- |
- |
| 14 |
Hb_013420_050 |
0.1990937921 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 15 |
Hb_042463_070 |
0.2001361245 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 16 |
Hb_006356_030 |
0.2022226416 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_009888_010 |
0.2025813348 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000923_110 |
0.2030483916 |
- |
- |
PREDICTED: uncharacterized protein LOC102624489 [Citrus sinensis] |
| 19 |
Hb_000100_120 |
0.2044807535 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 20 |
Hb_001383_080 |
0.2055554115 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |