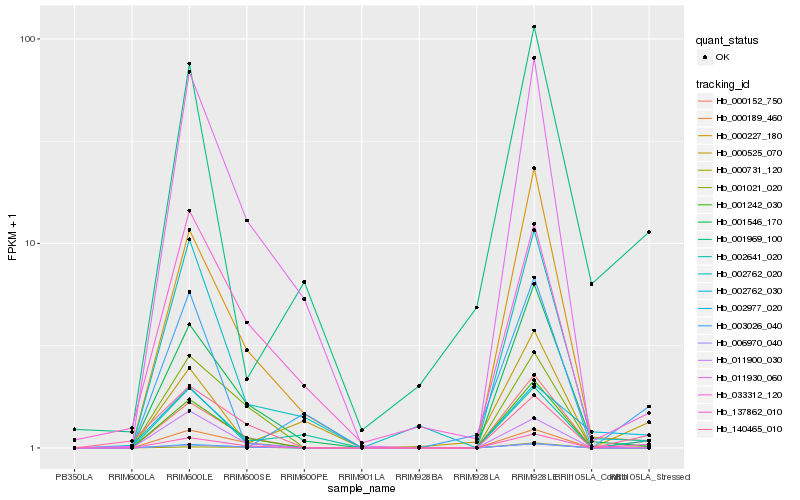
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137862_010 |
0.0 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000152_750 |
0.1417905897 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 3 |
Hb_006970_040 |
0.1548067029 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 4 |
Hb_140465_010 |
0.2051649533 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 5 |
Hb_000731_120 |
0.2200412244 |
- |
- |
Beta-glucosidase, putative [Ricinus communis] |
| 6 |
Hb_002641_020 |
0.2406458058 |
- |
- |
3'-5' exonuclease, putative [Ricinus communis] |
| 7 |
Hb_001242_030 |
0.2497514857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002762_030 |
0.2510638369 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 9 |
Hb_002977_020 |
0.2534238479 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 10 |
Hb_001546_170 |
0.2542273553 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000525_070 |
0.2604521741 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like [Jatropha curcas] |
| 12 |
Hb_000189_460 |
0.2607640132 |
- |
- |
- |
| 13 |
Hb_000227_180 |
0.2618684483 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001969_100 |
0.2662545932 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 15 |
Hb_003026_040 |
0.2667653616 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 16 |
Hb_001021_020 |
0.2670211417 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 17 |
Hb_033312_120 |
0.2721406111 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_011930_060 |
0.2733572643 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_002762_020 |
0.2757427087 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 20 |
Hb_011900_030 |
0.2765011179 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |