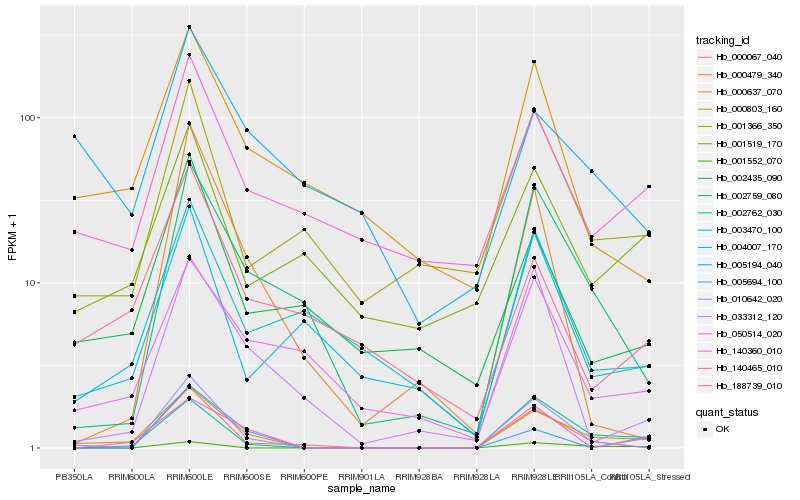
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_25955 [Jatropha curcas] |
| 2 |
Hb_002762_030 |
0.2234357051 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 3 |
Hb_010642_020 |
0.2288755174 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 4 |
Hb_140465_010 |
0.2318112971 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 5 |
Hb_002759_080 |
0.2475059653 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 6 |
Hb_140360_010 |
0.2662305237 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_000637_070 |
0.2685659912 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 8 |
Hb_050514_020 |
0.2719460715 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 9 |
Hb_000067_040 |
0.2745573903 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 10 |
Hb_033312_120 |
0.2759426318 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001366_350 |
0.2774191556 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 12 |
Hb_002435_090 |
0.27941597 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003470_100 |
0.2866156881 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_004007_170 |
0.2877452697 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_001519_170 |
0.2891245597 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005194_040 |
0.2892360701 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_005694_100 |
0.2901388417 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 18 |
Hb_188739_010 |
0.2906826174 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000479_340 |
0.2914867013 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 20 |
Hb_001552_070 |
0.2938972954 |
- |
- |
EXS family protein [Populus trichocarpa] |