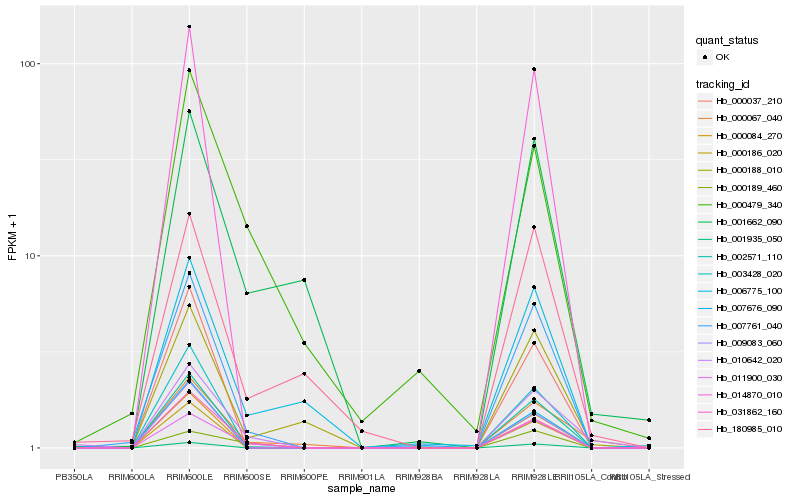
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010642_020 |
0.0 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 2 |
Hb_000067_040 |
0.1355045769 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_031862_160 |
0.1608054827 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_011900_030 |
0.1612408562 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 5 |
Hb_007761_040 |
0.1808365007 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 6 |
Hb_002571_110 |
0.1972300576 |
- |
- |
PREDICTED: phospholipase A1-II 1-like [Jatropha curcas] |
| 7 |
Hb_000479_340 |
0.2086551057 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 8 |
Hb_000188_010 |
0.211629196 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 9 |
Hb_180985_010 |
0.2155514508 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000189_460 |
0.21682008 |
- |
- |
- |
| 11 |
Hb_014870_010 |
0.2179901795 |
- |
- |
PREDICTED: uncharacterized protein LOC104891359 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000084_270 |
0.2183426943 |
- |
- |
hypothetical protein POPTR_0006s22820g [Populus trichocarpa] |
| 13 |
Hb_000186_020 |
0.2189201377 |
- |
- |
PREDICTED: uncharacterized protein LOC104901327 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_001662_090 |
0.2194713067 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001935_050 |
0.2219714531 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 [Jatropha curcas] |
| 16 |
Hb_007676_090 |
0.2237406695 |
- |
- |
PREDICTED: uncharacterized protein LOC105644038 [Jatropha curcas] |
| 17 |
Hb_006775_100 |
0.2238454995 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_003428_020 |
0.2247551963 |
- |
- |
PREDICTED: uncharacterized protein LOC105140247 [Populus euphratica] |
| 19 |
Hb_000037_210 |
0.2253938532 |
- |
- |
- |
| 20 |
Hb_009083_060 |
0.2256080217 |
- |
- |
PREDICTED: uncharacterized protein LOC103344557 [Prunus mume] |