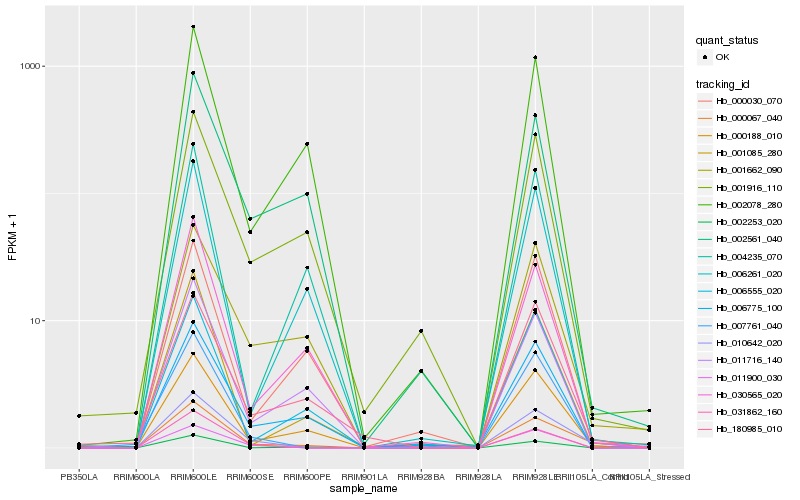
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000067_040 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 2 |
Hb_010642_020 |
0.1355045769 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 3 |
Hb_000188_010 |
0.1418748469 |
- |
- |
PREDICTED: cytochrome P450 94C1-like [Jatropha curcas] |
| 4 |
Hb_001662_090 |
0.164630084 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_180985_010 |
0.1709383178 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_006775_100 |
0.174824625 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002253_020 |
0.1756999473 |
- |
- |
PREDICTED: callose synthase 5 [Jatropha curcas] |
| 8 |
Hb_011716_140 |
0.1757531862 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 9 |
Hb_030565_020 |
0.1777552197 |
- |
- |
PREDICTED: protein ECERIFERUM 26 [Jatropha curcas] |
| 10 |
Hb_002561_040 |
0.1787690137 |
- |
- |
Photosynthetic electron transfer C [Theobroma cacao] |
| 11 |
Hb_001916_110 |
0.1840736444 |
- |
- |
PREDICTED: uncharacterized protein LOC105641510 [Jatropha curcas] |
| 12 |
Hb_002078_280 |
0.1845213833 |
- |
- |
chlorophyll a/b-binding protein type II precursor [Populus trichocarpa] |
| 13 |
Hb_031862_160 |
0.1850809014 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_001085_280 |
0.185194676 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 15 |
Hb_006261_020 |
0.191515773 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 16 |
Hb_011900_030 |
0.1938300889 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 17 |
Hb_006555_020 |
0.1944036242 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 18 |
Hb_000030_070 |
0.1947518245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007761_040 |
0.1951008317 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |
| 20 |
Hb_004235_070 |
0.1975707778 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |