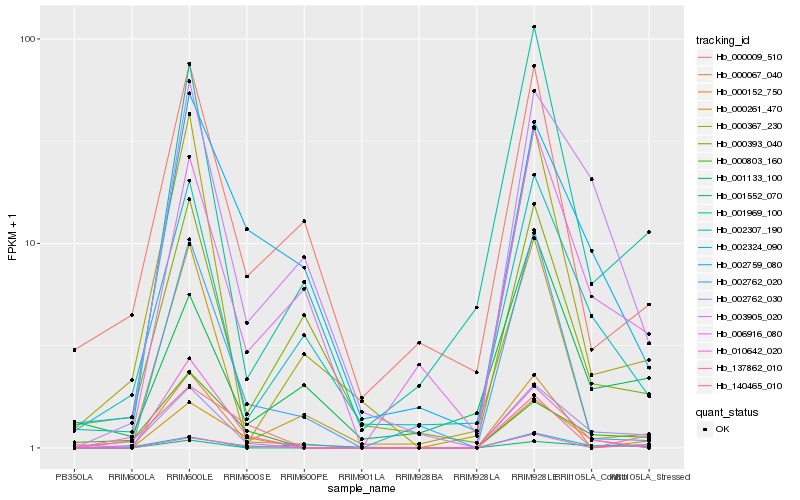
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002762_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 2 |
Hb_001552_070 |
0.1476184957 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 3 |
Hb_003905_020 |
0.2182978743 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 4 |
Hb_000803_160 |
0.2234357051 |
- |
- |
hypothetical protein JCGZ_25955 [Jatropha curcas] |
| 5 |
Hb_001969_100 |
0.2236665118 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 6 |
Hb_002307_190 |
0.2271340477 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 7 |
Hb_006916_080 |
0.23290718 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 8 |
Hb_000367_230 |
0.2374334028 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002324_090 |
0.2469655611 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 10 |
Hb_137862_010 |
0.2510638369 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000152_750 |
0.2613286313 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 12 |
Hb_000067_040 |
0.2645656563 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_002759_080 |
0.2676287547 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 14 |
Hb_010642_020 |
0.2679079229 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 15 |
Hb_000261_470 |
0.2723933498 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 16 |
Hb_000393_040 |
0.2750397372 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 17 |
Hb_002762_020 |
0.2764028998 |
- |
- |
hypothetical protein VITISV_043202 [Vitis vinifera] |
| 18 |
Hb_001133_100 |
0.2781774846 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 19 |
Hb_140465_010 |
0.2802663879 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 20 |
Hb_000009_510 |
0.2842236267 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |