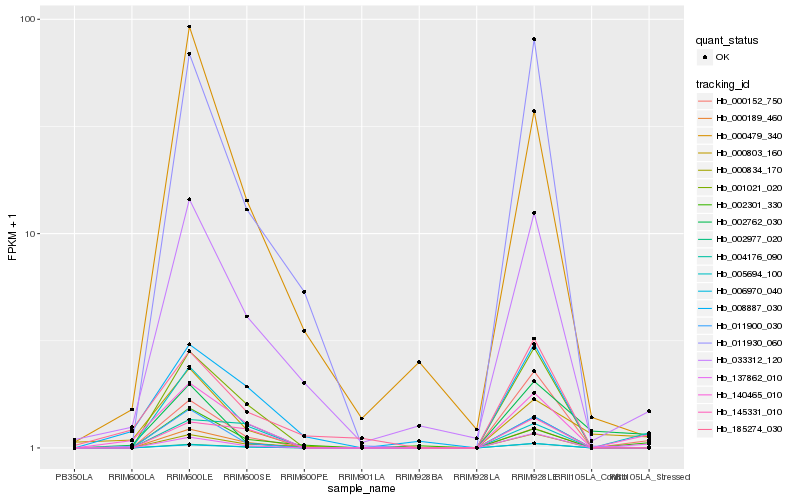
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140465_010 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 2 |
Hb_137862_010 |
0.2051649533 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_006970_040 |
0.2081101288 |
- |
- |
PREDICTED: uncharacterized protein LOC105650489 [Jatropha curcas] |
| 4 |
Hb_033312_120 |
0.215095148 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000803_160 |
0.2318112971 |
- |
- |
hypothetical protein JCGZ_25955 [Jatropha curcas] |
| 6 |
Hb_001021_020 |
0.2337819274 |
- |
- |
hypothetical protein JCGZ_13899 [Jatropha curcas] |
| 7 |
Hb_000189_460 |
0.235273872 |
- |
- |
- |
| 8 |
Hb_008887_030 |
0.2353183621 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 9 |
Hb_002301_330 |
0.2429652053 |
- |
- |
PREDICTED: late embryogenesis abundant protein, group 3-like [Jatropha curcas] |
| 10 |
Hb_002977_020 |
0.2456658676 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 1.8-like [Nelumbo nucifera] |
| 11 |
Hb_000152_750 |
0.2466287638 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 12 |
Hb_005694_100 |
0.2576636337 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 13 |
Hb_145331_010 |
0.2610466089 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0016s07690g [Populus trichocarpa] |
| 14 |
Hb_011900_030 |
0.261949131 |
- |
- |
hypothetical protein JCGZ_13686 [Jatropha curcas] |
| 15 |
Hb_185274_030 |
0.2620071315 |
- |
- |
hypothetical protein JCGZ_15170 [Jatropha curcas] |
| 16 |
Hb_004176_090 |
0.2747846861 |
- |
- |
Nuc-1 negative regulatory protein preg, putative [Ricinus communis] |
| 17 |
Hb_011930_060 |
0.2781434247 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_000834_170 |
0.2795438155 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 19 |
Hb_002762_030 |
0.2802663879 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 20 |
Hb_000479_340 |
0.2805673387 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |