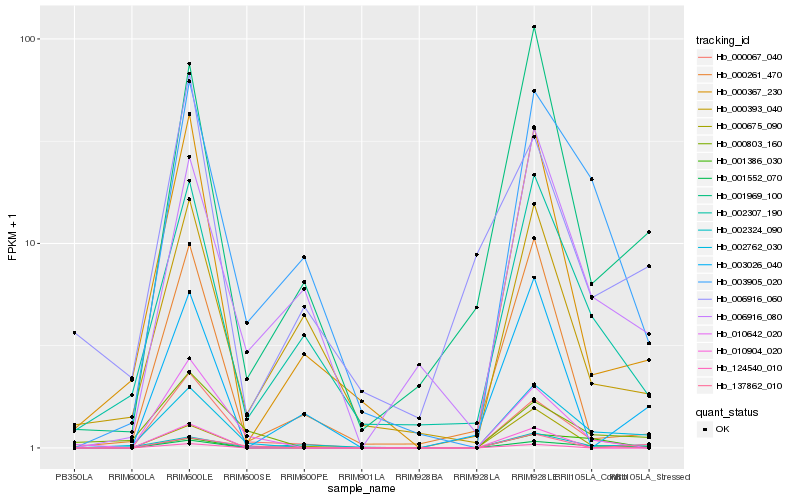
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001552_070 |
0.0 |
- |
- |
EXS family protein [Populus trichocarpa] |
| 2 |
Hb_002762_030 |
0.1476184957 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 3 |
Hb_003905_020 |
0.2450834559 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 4 |
Hb_001969_100 |
0.2603545457 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 5 |
Hb_000367_230 |
0.2695691628 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002307_190 |
0.273368272 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 7 |
Hb_006916_080 |
0.2846527662 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 8 |
Hb_001386_030 |
0.2881272418 |
- |
- |
PREDICTED: peroxidase 55 [Jatropha curcas] |
| 9 |
Hb_000803_160 |
0.2938972954 |
- |
- |
hypothetical protein JCGZ_25955 [Jatropha curcas] |
| 10 |
Hb_000067_040 |
0.2970568635 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 11 |
Hb_003026_040 |
0.3038097584 |
- |
- |
Uncharacterized protein TCM_007861 [Theobroma cacao] |
| 12 |
Hb_002324_090 |
0.3067486254 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_006916_060 |
0.309447891 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 14 |
Hb_010642_020 |
0.3130692902 |
- |
- |
NDH-dependent cyclic electron flow 1 isoform 2 [Theobroma cacao] |
| 15 |
Hb_000261_470 |
0.3132546152 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FAMA isoform X2 [Populus euphratica] |
| 16 |
Hb_137862_010 |
0.314687864 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000675_090 |
0.3168175819 |
- |
- |
hypothetical protein SORBIDRAFT_03g028570 [Sorghum bicolor] |
| 18 |
Hb_000393_040 |
0.3208651831 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 19 |
Hb_124540_010 |
0.3254518235 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1-like [Populus euphratica] |
| 20 |
Hb_010904_020 |
0.3254629451 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 [Populus euphratica] |