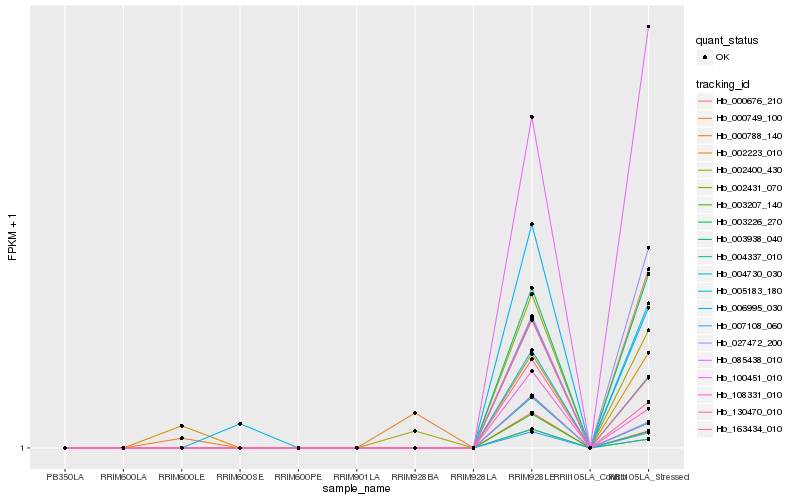
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027472_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_100451_010 |
0.0330672856 |
- |
- |
- |
| 3 |
Hb_003938_040 |
0.077843991 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 4 |
Hb_007108_060 |
0.1114165821 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_006995_030 |
0.2057037668 |
- |
- |
PREDICTED: uncharacterized protein LOC102663993 [Glycine max] |
| 6 |
Hb_005183_180 |
0.2101874026 |
- |
- |
alpha amylase family protein [Populus trichocarpa] |
| 7 |
Hb_002400_430 |
0.2284589017 |
- |
- |
- |
| 8 |
Hb_003226_270 |
0.2292192638 |
- |
- |
hypothetical protein POPTR_0014s12390g [Populus trichocarpa] |
| 9 |
Hb_130470_010 |
0.2296594418 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 10 |
Hb_163434_010 |
0.2386870616 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 11 |
Hb_108331_010 |
0.2411927945 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 12 |
Hb_000788_140 |
0.2458373991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_085438_010 |
0.2461805942 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_002431_070 |
0.2462708862 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 15 |
Hb_004730_030 |
0.2465315596 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 16 |
Hb_000749_100 |
0.2490560467 |
- |
- |
- |
| 17 |
Hb_000676_210 |
0.249498369 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 18 |
Hb_003207_140 |
0.2497847665 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 19 |
Hb_004337_010 |
0.2526032344 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_002223_010 |
0.2526176886 |
- |
- |
hypothetical protein VITISV_009489 [Vitis vinifera] |