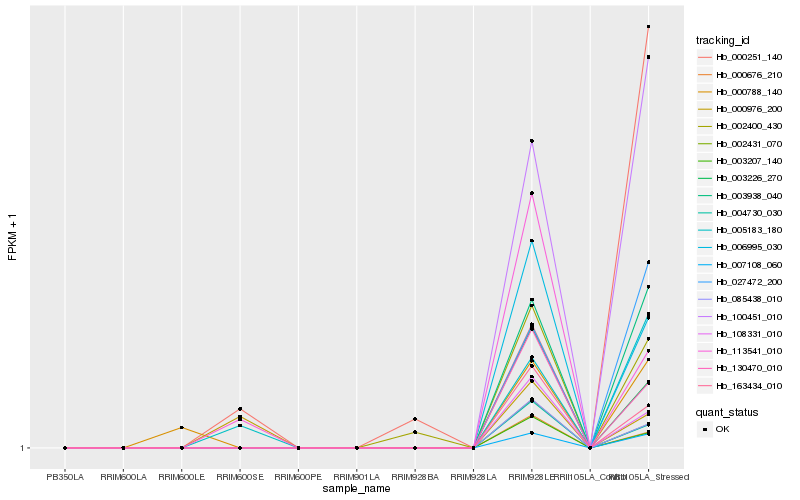
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_180 |
0.0 |
- |
- |
alpha amylase family protein [Populus trichocarpa] |
| 2 |
Hb_027472_200 |
0.2101874026 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_100451_010 |
0.2114196438 |
- |
- |
- |
| 4 |
Hb_003938_040 |
0.2207542293 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 5 |
Hb_007108_060 |
0.2329643886 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_000251_140 |
0.2366676307 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 7 |
Hb_000976_200 |
0.2764486941 |
- |
- |
- |
| 8 |
Hb_006995_030 |
0.2852863669 |
- |
- |
PREDICTED: uncharacterized protein LOC102663993 [Glycine max] |
| 9 |
Hb_113541_010 |
0.2935483245 |
- |
- |
PREDICTED: uncharacterized protein LOC105762713 [Gossypium raimondii] |
| 10 |
Hb_003226_270 |
0.3013569858 |
- |
- |
hypothetical protein POPTR_0014s12390g [Populus trichocarpa] |
| 11 |
Hb_130470_010 |
0.301666516 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 12 |
Hb_002400_430 |
0.3018906251 |
- |
- |
- |
| 13 |
Hb_163434_010 |
0.3080801694 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 14 |
Hb_108331_010 |
0.3098819171 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 15 |
Hb_085438_010 |
0.3134952559 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 16 |
Hb_002431_070 |
0.3135609906 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA11 [Jatropha curcas] |
| 17 |
Hb_004730_030 |
0.3137508307 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 18 |
Hb_000788_140 |
0.3155783989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000676_210 |
0.3159180992 |
- |
- |
Cytochrome P450 superfamily protein [Theobroma cacao] |
| 20 |
Hb_003207_140 |
0.3161279546 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |