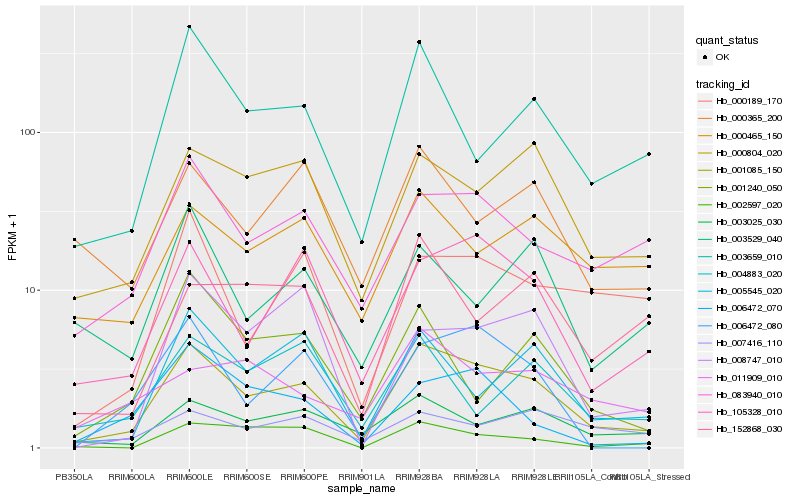
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_150 |
0.0 |
- |
- |
Purple acid phosphatase 15 [Theobroma cacao] |
| 2 |
Hb_105328_010 |
0.1759504267 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 3 |
Hb_000804_020 |
0.1846566349 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_083940_010 |
0.1849259826 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 5 |
Hb_003025_030 |
0.1883354056 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 6 |
Hb_003659_010 |
0.1887046336 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 7 |
Hb_005545_020 |
0.1988159743 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 8 |
Hb_000465_150 |
0.2106426623 |
- |
- |
PREDICTED: peroxisomal acyl-coenzyme A oxidase 1 [Jatropha curcas] |
| 9 |
Hb_000189_170 |
0.2116254024 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_002597_020 |
0.21237687 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 11 |
Hb_006472_080 |
0.2128645364 |
- |
- |
- |
| 12 |
Hb_000365_200 |
0.2135744943 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 13 |
Hb_007416_110 |
0.2197452674 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_152868_030 |
0.2223557173 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 15 |
Hb_003529_040 |
0.2244948865 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_006472_070 |
0.2251133495 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 17 |
Hb_004883_020 |
0.2266872632 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 18 |
Hb_001240_050 |
0.2274725729 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 19 |
Hb_008747_010 |
0.2276162869 |
- |
- |
PREDICTED: protein argonaute 2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_011909_010 |
0.2276247575 |
- |
- |
PREDICTED: uncharacterized protein LOC100246941 [Vitis vinifera] |