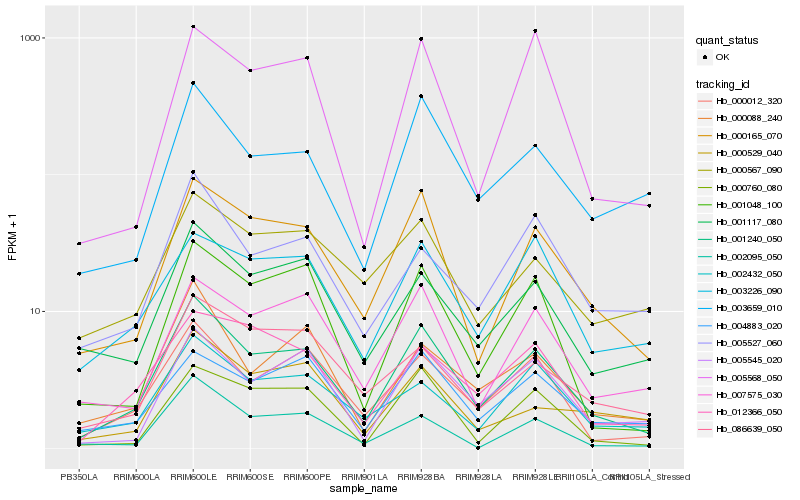
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001240_050 |
0.0 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 2 |
Hb_000165_070 |
0.1062929935 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 3 |
Hb_000012_320 |
0.1228562753 |
- |
- |
PREDICTED: uncharacterized protein LOC105638164 [Jatropha curcas] |
| 4 |
Hb_001048_100 |
0.1405273955 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 5 |
Hb_003659_010 |
0.1416935116 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 6 |
Hb_005545_020 |
0.1424835375 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 7 |
Hb_001117_080 |
0.1425571645 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007575_030 |
0.1433596017 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 9 |
Hb_000088_240 |
0.1452980491 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000760_080 |
0.1479076944 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 11 |
Hb_002432_050 |
0.1486088809 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 12 |
Hb_012366_050 |
0.1525224228 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 13 |
Hb_086639_050 |
0.1530149968 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004883_020 |
0.1544871112 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 15 |
Hb_005527_060 |
0.1592645801 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 16 |
Hb_000529_040 |
0.1602413894 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 17 |
Hb_005568_050 |
0.1621551857 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 18 |
Hb_000567_090 |
0.1629538051 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_002095_050 |
0.1631575178 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 20 |
Hb_003226_090 |
0.1640737439 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |