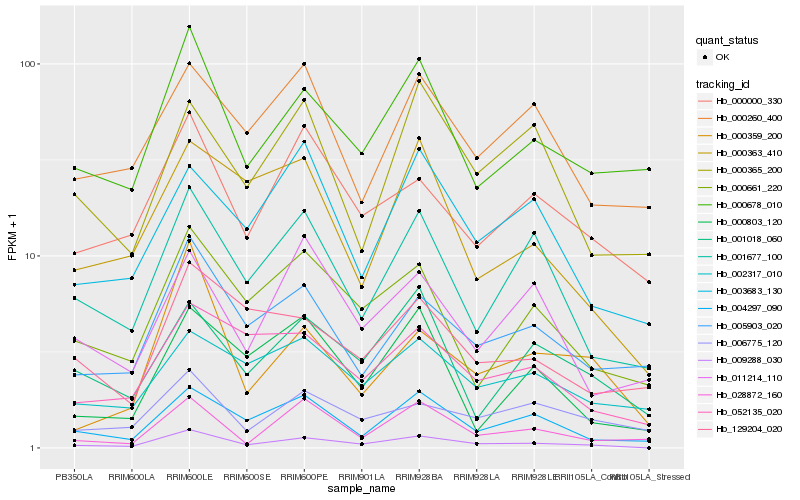
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009288_030 |
0.0 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 2 |
Hb_004297_090 |
0.1637631995 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 3 |
Hb_001677_100 |
0.1717858934 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |
| 4 |
Hb_000678_010 |
0.1731558032 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000661_220 |
0.1793755798 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 55-like [Jatropha curcas] |
| 6 |
Hb_005903_020 |
0.1803652191 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 7 |
Hb_000000_330 |
0.1830037268 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 8 |
Hb_052135_020 |
0.1834484417 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 9 |
Hb_000359_200 |
0.1852876661 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 10 |
Hb_000363_410 |
0.1906116275 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 11 |
Hb_006775_120 |
0.1909762135 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 12 |
Hb_028872_160 |
0.1915185686 |
- |
- |
PREDICTED: ATP-dependent DNA helicase Q-like 5 [Jatropha curcas] |
| 13 |
Hb_001018_060 |
0.1919508552 |
- |
- |
hypothetical protein B456_001G028000 [Gossypium raimondii] |
| 14 |
Hb_003683_130 |
0.1947657496 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002317_010 |
0.1968785175 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000803_120 |
0.2006617145 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011214_110 |
0.2024173841 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 18 |
Hb_000260_400 |
0.2029052906 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 19 |
Hb_000365_200 |
0.2029545199 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 20 |
Hb_129204_020 |
0.2031650944 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |