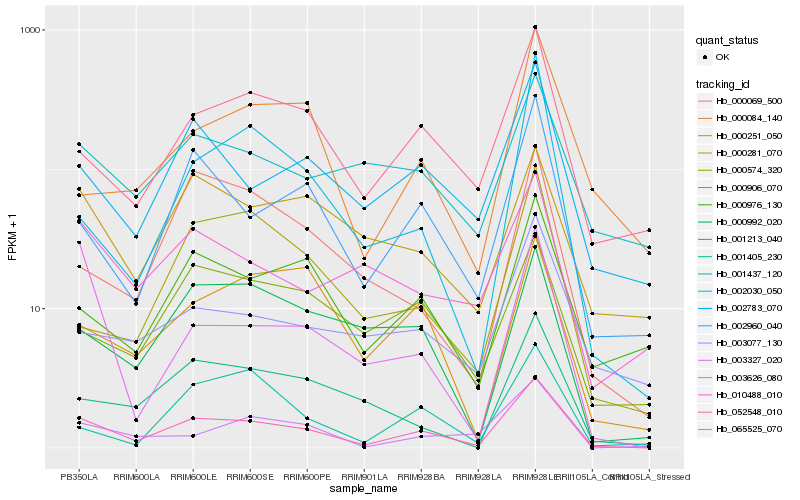
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052548_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 2 |
Hb_002960_040 |
0.1725087095 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 3 |
Hb_065525_070 |
0.1802462668 |
- |
- |
unnamed protein product [Homo sapiens] |
| 4 |
Hb_002783_070 |
0.1852119462 |
- |
- |
PREDICTED: endoplasmin homolog [Jatropha curcas] |
| 5 |
Hb_000976_130 |
0.1951593699 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 6 |
Hb_002030_050 |
0.2014451096 |
- |
- |
classical arabinogalactan protein 7 precursor [Jatropha curcas] |
| 7 |
Hb_003327_020 |
0.2051776154 |
- |
- |
PREDICTED: phytosulfokine receptor 2-like, partial [Jatropha curcas] |
| 8 |
Hb_001213_040 |
0.2102406792 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 9 |
Hb_000992_020 |
0.2137120128 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 10 |
Hb_000574_320 |
0.2163253305 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 11 |
Hb_000906_070 |
0.2193393891 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003077_130 |
0.2196666763 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 13 |
Hb_000281_070 |
0.220075473 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 14 |
Hb_001437_120 |
0.2203493275 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 15 |
Hb_001405_230 |
0.2217672622 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 16 |
Hb_000084_140 |
0.2224623545 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 17 |
Hb_010488_010 |
0.2226138696 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000251_050 |
0.2237210243 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 19 |
Hb_003626_080 |
0.2251765824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000069_500 |
0.2255742546 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |