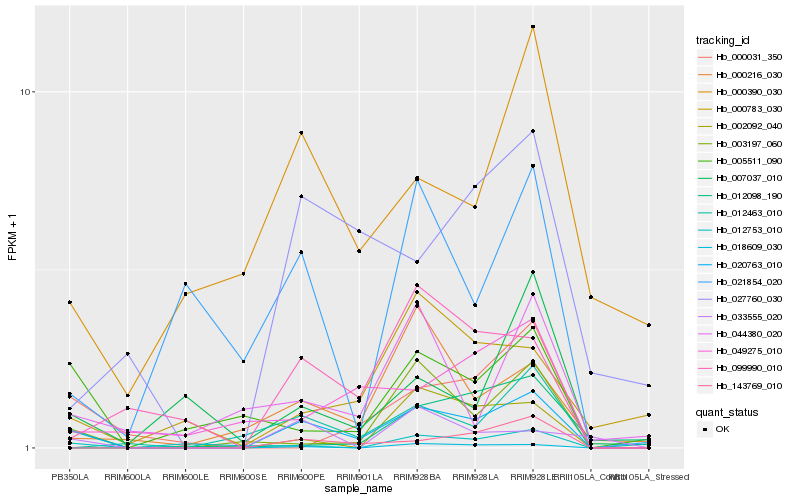
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020763_010 |
0.0 |
- |
- |
polyamine oxidase, putative [Ricinus communis] |
| 2 |
Hb_003197_060 |
0.2833372533 |
- |
- |
hypothetical protein POPTR_0015s11900g [Populus trichocarpa] |
| 3 |
Hb_143769_010 |
0.287410187 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 4 |
Hb_018609_030 |
0.3033531445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005511_090 |
0.3099784664 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |
| 6 |
Hb_099990_010 |
0.3104040329 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 11 [Vitis vinifera] |
| 7 |
Hb_000216_030 |
0.3152371576 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 8 |
Hb_012463_010 |
0.3169207506 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_012753_010 |
0.3212228201 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_000031_350 |
0.3250299533 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic isoform X2 [Cucumis melo] |
| 11 |
Hb_007037_010 |
0.3254744797 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 12 |
Hb_012098_190 |
0.3265957696 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_027760_030 |
0.3270870857 |
- |
- |
- |
| 14 |
Hb_049275_010 |
0.3335495595 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 15 |
Hb_000783_030 |
0.3363770482 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 16 |
Hb_044380_020 |
0.3382576358 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_002092_040 |
0.3406323142 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 18 |
Hb_000390_030 |
0.3408765754 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 19 |
Hb_021854_020 |
0.3413398865 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 20 |
Hb_033555_020 |
0.3424490253 |
- |
- |
Uncharacterized protein TCM_003129 [Theobroma cacao] |