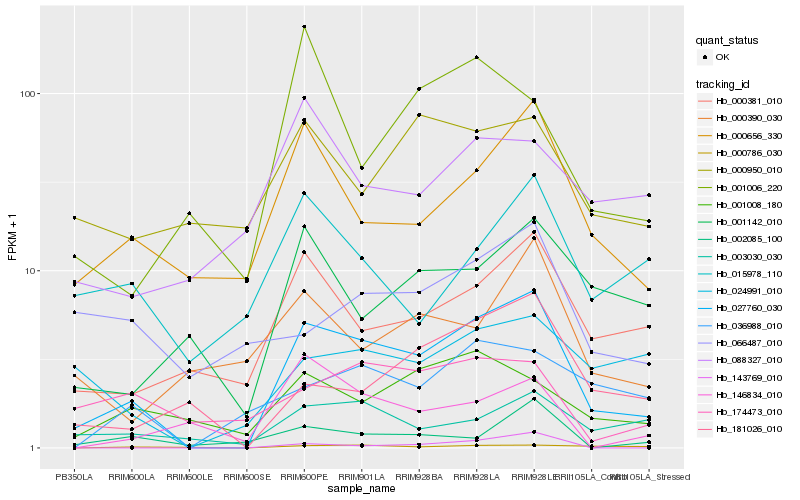
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027760_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000656_330 |
0.2351646676 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_000381_010 |
0.2425385264 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_003030_030 |
0.2579651238 |
- |
- |
- |
| 5 |
Hb_181026_010 |
0.2586742615 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_001008_180 |
0.2691102041 |
- |
- |
- |
| 7 |
Hb_001142_010 |
0.2710819979 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 8 |
Hb_036988_010 |
0.2727549218 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 9 |
Hb_000786_030 |
0.2768914448 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 10 |
Hb_024991_010 |
0.2799046615 |
- |
- |
hypothetical protein B456_007G217100 [Gossypium raimondii] |
| 11 |
Hb_143769_010 |
0.2858438425 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 12 |
Hb_174473_010 |
0.2863437057 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 13 |
Hb_002085_100 |
0.2886944409 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000390_030 |
0.2890366874 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 15 |
Hb_015978_110 |
0.2892991391 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62470, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_000950_010 |
0.2897612904 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 17 |
Hb_088327_010 |
0.2921694594 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 18 |
Hb_066487_010 |
0.2935121217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_146834_010 |
0.2935813836 |
- |
- |
Uncharacterized protein TCM_035385 [Theobroma cacao] |
| 20 |
Hb_001006_220 |
0.2942044956 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |