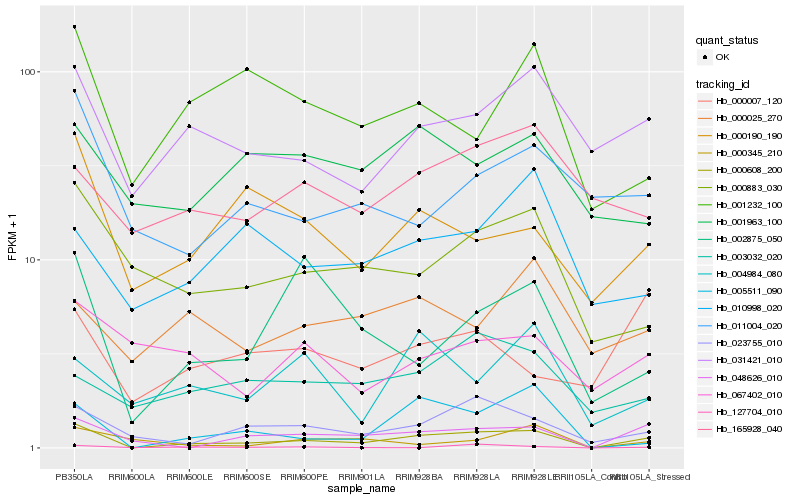
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_200 |
0.0 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Pyrus x bretschneideri] |
| 2 |
Hb_048626_010 |
0.2396872802 |
- |
- |
- |
| 3 |
Hb_023755_010 |
0.2428494856 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 4 |
Hb_005511_090 |
0.2430800944 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |
| 5 |
Hb_031421_010 |
0.2619047442 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 6 |
Hb_000883_030 |
0.2720421185 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 7 |
Hb_002875_050 |
0.2760657597 |
- |
- |
PREDICTED: proteoglycan 4 [Jatropha curcas] |
| 8 |
Hb_127704_010 |
0.2801377935 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000190_190 |
0.2827106619 |
- |
- |
PREDICTED: BI1-like protein [Gossypium raimondii] |
| 10 |
Hb_001232_100 |
0.2889736142 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 11 |
Hb_003032_020 |
0.2893204677 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55740, chloroplastic [Jatropha curcas] |
| 12 |
Hb_011004_020 |
0.2895540572 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 13 |
Hb_004984_080 |
0.2932244013 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_010998_020 |
0.2967540924 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |
| 15 |
Hb_165928_040 |
0.2993006978 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 16 |
Hb_000025_270 |
0.3031787106 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_001963_100 |
0.3035701103 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 18 |
Hb_000007_120 |
0.3051478291 |
desease resistance |
Gene Name: PRK |
uridine cytidine kinase I, putative [Ricinus communis] |
| 19 |
Hb_000345_210 |
0.3071134867 |
- |
- |
PREDICTED: uncharacterized protein LOC105647523 [Jatropha curcas] |
| 20 |
Hb_067402_010 |
0.3079177061 |
- |
- |
hypothetical protein CISIN_1g0029712mg, partial [Citrus sinensis] |