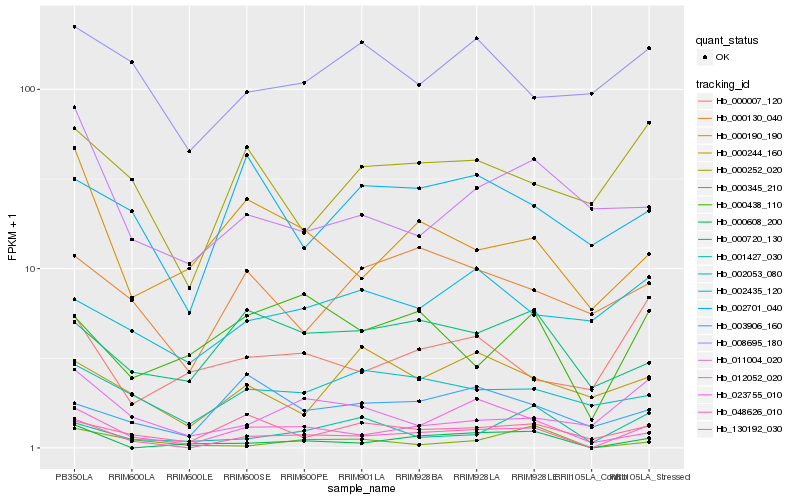
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048626_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_023755_010 |
0.2237124653 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_000608_200 |
0.2396872802 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Pyrus x bretschneideri] |
| 4 |
Hb_000252_020 |
0.2499878264 |
- |
- |
PREDICTED: alpha-glucan water dikinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_130192_030 |
0.25981612 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 6 |
Hb_012052_020 |
0.2639911072 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 7 |
Hb_001427_030 |
0.2679852961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000007_120 |
0.2682068414 |
desease resistance |
Gene Name: PRK |
uridine cytidine kinase I, putative [Ricinus communis] |
| 9 |
Hb_000438_110 |
0.2690033598 |
- |
- |
PREDICTED: adenine/guanine permease AZG1 [Jatropha curcas] |
| 10 |
Hb_002053_080 |
0.2691044289 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21300 [Jatropha curcas] |
| 11 |
Hb_000130_040 |
0.2691158889 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 12 |
Hb_000720_130 |
0.2721140531 |
- |
- |
hypothetical protein PRUPE_ppa012392mg [Prunus persica] |
| 13 |
Hb_008695_180 |
0.2785519331 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 14 |
Hb_002435_120 |
0.2798450445 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000244_160 |
0.2799235255 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g24000, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_003906_160 |
0.2809496824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_002701_040 |
0.281062366 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 18 |
Hb_011004_020 |
0.2812001931 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 19 |
Hb_000190_190 |
0.28180453 |
- |
- |
PREDICTED: BI1-like protein [Gossypium raimondii] |
| 20 |
Hb_000345_210 |
0.2818612963 |
- |
- |
PREDICTED: uncharacterized protein LOC105647523 [Jatropha curcas] |