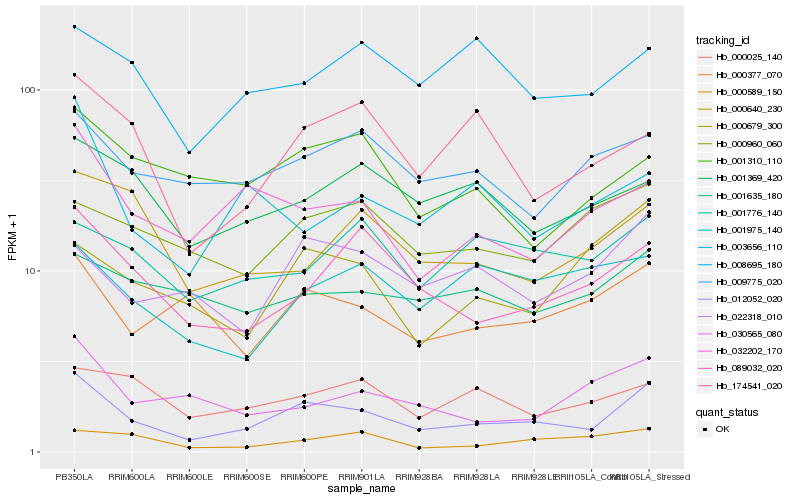
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012052_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 2 |
Hb_089032_020 |
0.1679461689 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 3 |
Hb_032202_170 |
0.1773471954 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 4 |
Hb_000377_070 |
0.1788820975 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 2 [Jatropha curcas] |
| 5 |
Hb_022318_010 |
0.1795611579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009775_020 |
0.1833300351 |
- |
- |
PREDICTED: vesicle transport protein GOT1B-like [Jatropha curcas] |
| 7 |
Hb_174541_020 |
0.1862551011 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 6-like [Populus euphratica] |
| 8 |
Hb_000589_150 |
0.1874535176 |
- |
- |
PREDICTED: factor of DNA methylation 1-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000679_300 |
0.1888001938 |
- |
- |
PREDICTED: uncharacterized protein LOC105637628 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000640_230 |
0.1903095803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001369_420 |
0.1908950375 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 12 |
Hb_030565_080 |
0.1910231946 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 13 |
Hb_000025_140 |
0.1911044627 |
- |
- |
las1-like family protein [Populus trichocarpa] |
| 14 |
Hb_001310_110 |
0.1922149818 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_003656_110 |
0.1927177449 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_008695_180 |
0.1935249748 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 17 |
Hb_000960_060 |
0.1940003043 |
- |
- |
PREDICTED: ADP-ribosylation factor 1-like 2 [Jatropha curcas] |
| 18 |
Hb_001776_140 |
0.1949300342 |
- |
- |
PREDICTED: probable GTP-binding protein OBGM, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_001635_180 |
0.1955559522 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At5g53840-like [Jatropha curcas] |
| 20 |
Hb_001975_140 |
0.1960693504 |
- |
- |
PREDICTED: urease accessory protein F [Jatropha curcas] |