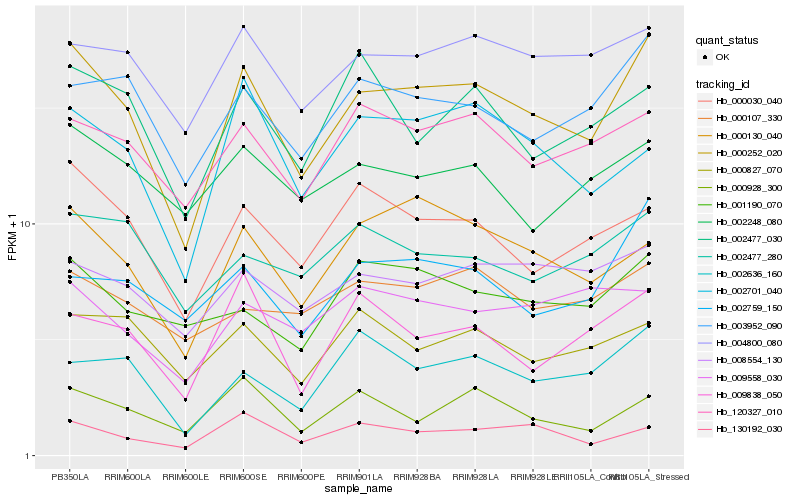
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_020 |
0.0 |
- |
- |
PREDICTED: alpha-glucan water dikinase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003952_090 |
0.1145894633 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000030_040 |
0.1172393457 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000130_040 |
0.1192063677 |
- |
- |
PREDICTED: dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 5 |
Hb_004800_080 |
0.1220659514 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 6 |
Hb_120327_010 |
0.1224716758 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 7 |
Hb_008554_130 |
0.1288522347 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105634317 [Jatropha curcas] |
| 8 |
Hb_002636_160 |
0.1289312615 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g13630 [Jatropha curcas] |
| 9 |
Hb_130192_030 |
0.1311240557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 10 |
Hb_002759_150 |
0.1315485544 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002248_080 |
0.1334789606 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002701_040 |
0.1339164062 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 13 |
Hb_009838_050 |
0.1342688783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002477_280 |
0.1347131395 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-2 [Jatropha curcas] |
| 15 |
Hb_000827_070 |
0.1347520553 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_001190_070 |
0.1349221281 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 17 |
Hb_002477_030 |
0.1358387493 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000928_300 |
0.136968454 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g03580 [Jatropha curcas] |
| 19 |
Hb_000107_330 |
0.1378720707 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38150 [Jatropha curcas] |
| 20 |
Hb_009558_030 |
0.1411541719 |
- |
- |
myb3r3, putative [Ricinus communis] |