| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
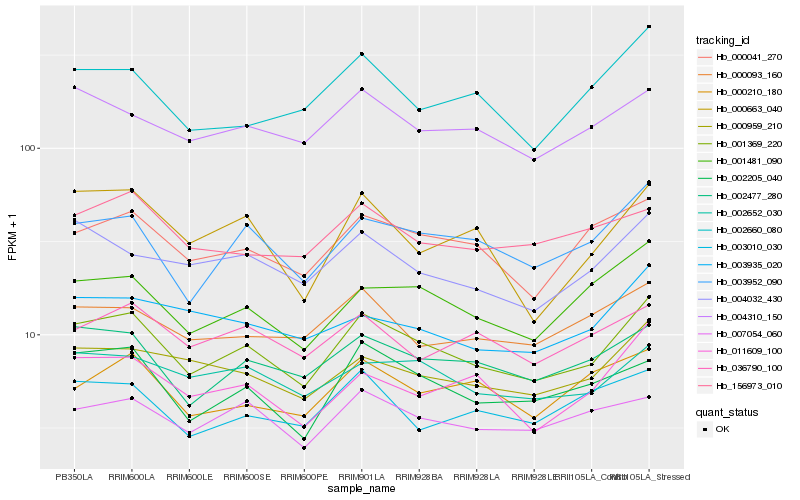
Hb_001369_220 |
0.0 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_003952_090 |
0.0734725248 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002477_280 |
0.0759827913 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-2 [Jatropha curcas] |
| 4 |
Hb_001481_090 |
0.0794357917 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 5 |
Hb_000041_270 |
0.0805731768 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 6 |
Hb_002205_040 |
0.0807084104 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000959_210 |
0.0832381879 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 8 |
Hb_002652_030 |
0.0834566194 |
- |
- |
PREDICTED: U3 small nucleolar RNA-interacting protein 2-like [Jatropha curcas] |
| 9 |
Hb_004310_150 |
0.088110372 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 10 |
Hb_004032_430 |
0.0883027956 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 11 |
Hb_007054_060 |
0.0896064203 |
- |
- |
- |
| 12 |
Hb_011609_100 |
0.0901647354 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000093_160 |
0.0935733782 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000663_040 |
0.0951348588 |
- |
- |
PREDICTED: uncharacterized protein LOC105650729 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002660_080 |
0.0954412359 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 16 |
Hb_003935_020 |
0.0968392953 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000210_180 |
0.0971344023 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 18 |
Hb_156973_010 |
0.0972872446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_036790_100 |
0.0976565516 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |
| 20 |
Hb_003010_030 |
0.0982970004 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |