| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
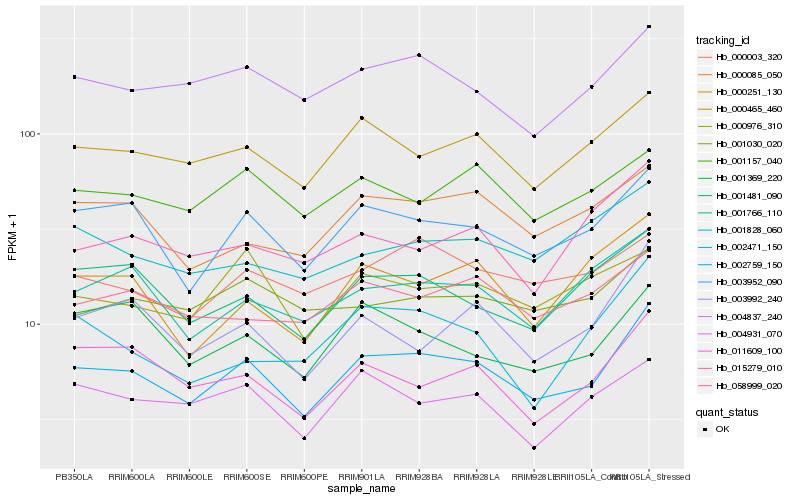
Hb_002759_150 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003952_090 |
0.0768807526 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000465_460 |
0.0895049398 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 4 |
Hb_004837_240 |
0.0970397873 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 5 |
Hb_001481_090 |
0.0984073863 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 6 |
Hb_001766_110 |
0.0997288856 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 7 |
Hb_003992_240 |
0.0998859635 |
- |
- |
- |
| 8 |
Hb_000085_050 |
0.1035650154 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_011609_100 |
0.1039351536 |
- |
- |
PREDICTED: 30S ribosomal protein 3, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000251_130 |
0.1056843566 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 11 |
Hb_004931_070 |
0.1081592483 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_058999_020 |
0.1083867706 |
- |
- |
PREDICTED: mitochondrial outer membrane import complex protein METAXIN [Jatropha curcas] |
| 13 |
Hb_000003_320 |
0.108503502 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 14 |
Hb_001157_040 |
0.1104834403 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 15 |
Hb_001369_220 |
0.1114353269 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 6B-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001828_060 |
0.1120818831 |
desease resistance |
Gene Name: AAA_31 |
nucleotide-binding protein, putative [Ricinus communis] |
| 17 |
Hb_001030_020 |
0.1120943323 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 18 |
Hb_015279_010 |
0.1121684011 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 19 |
Hb_002471_150 |
0.1124813165 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000976_310 |
0.1126724158 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |