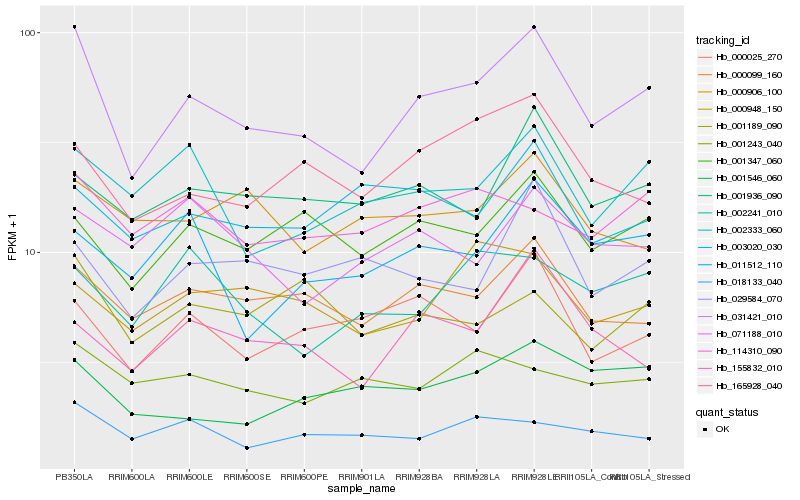
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031421_010 |
0.0 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 2 |
Hb_000099_160 |
0.1273347805 |
- |
- |
PREDICTED: potassium transporter 3 isoform X2 [Populus euphratica] |
| 3 |
Hb_001347_060 |
0.1372541097 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_002333_060 |
0.1413463748 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_165928_040 |
0.1428843975 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase component HRD3A [Jatropha curcas] |
| 6 |
Hb_001936_090 |
0.1457602843 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_270 |
0.146687416 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_114310_090 |
0.1506248871 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase [Jatropha curcas] |
| 9 |
Hb_018133_040 |
0.1507341709 |
- |
- |
PREDICTED: probable apyrase 7 [Jatropha curcas] |
| 10 |
Hb_155832_010 |
0.1510641794 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 11 |
Hb_011512_110 |
0.1522149571 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_029584_070 |
0.1538150421 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 13 |
Hb_001189_090 |
0.1545221436 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 14 |
Hb_001546_060 |
0.1553208935 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 15 |
Hb_071188_010 |
0.1559859151 |
- |
- |
hypothetical protein POPTR_0013s025001g, partial [Populus trichocarpa] |
| 16 |
Hb_002241_010 |
0.1571470672 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 17 |
Hb_000948_150 |
0.1572199655 |
- |
- |
PREDICTED: heme-binding-like protein At3g10130, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000906_100 |
0.1603514365 |
- |
- |
PREDICTED: uncharacterized protein LOC105648070 [Jatropha curcas] |
| 19 |
Hb_001243_040 |
0.1615905346 |
- |
- |
PREDICTED: uncharacterized protein LOC104104316 [Nicotiana tomentosiformis] |
| 20 |
Hb_003020_030 |
0.1617493664 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |