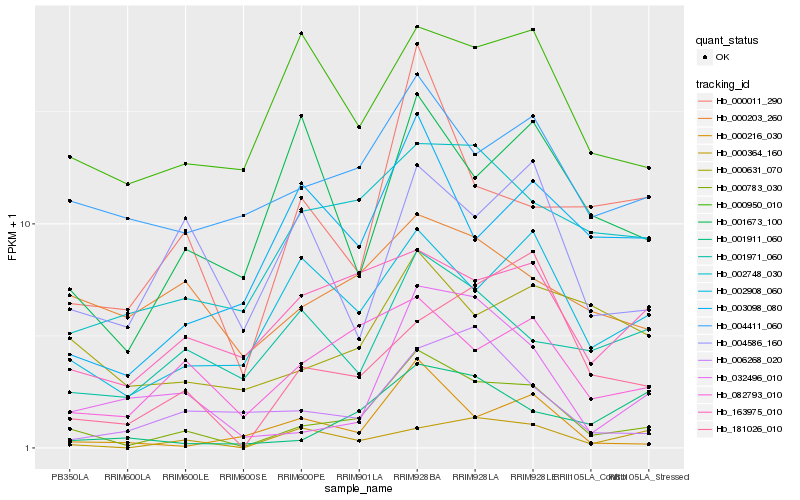
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000783_030 |
0.0 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 2 |
Hb_032496_010 |
0.2169183348 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 3 |
Hb_082793_010 |
0.2332745059 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_002748_030 |
0.2414734926 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 5 |
Hb_000011_290 |
0.2465677925 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 6 |
Hb_002908_060 |
0.2470906876 |
- |
- |
unnamed protein product, partial [Coffea canephora] |
| 7 |
Hb_000216_030 |
0.247271057 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 8 |
Hb_004411_060 |
0.2479187951 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 9 |
Hb_181026_010 |
0.2523953808 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_000631_070 |
0.2548368681 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 11 |
Hb_001911_060 |
0.2561222972 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_163975_010 |
0.2590053737 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 13 |
Hb_006268_020 |
0.2620397657 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 14 |
Hb_003098_080 |
0.2630283194 |
- |
- |
- |
| 15 |
Hb_001971_060 |
0.2659539297 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 16 |
Hb_000950_010 |
0.2683515143 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 17 |
Hb_000364_160 |
0.2694075091 |
- |
- |
PREDICTED: uncharacterized protein LOC105639754 [Jatropha curcas] |
| 18 |
Hb_000203_260 |
0.271305288 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 19 |
Hb_004586_160 |
0.271837938 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001673_100 |
0.2738271016 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |