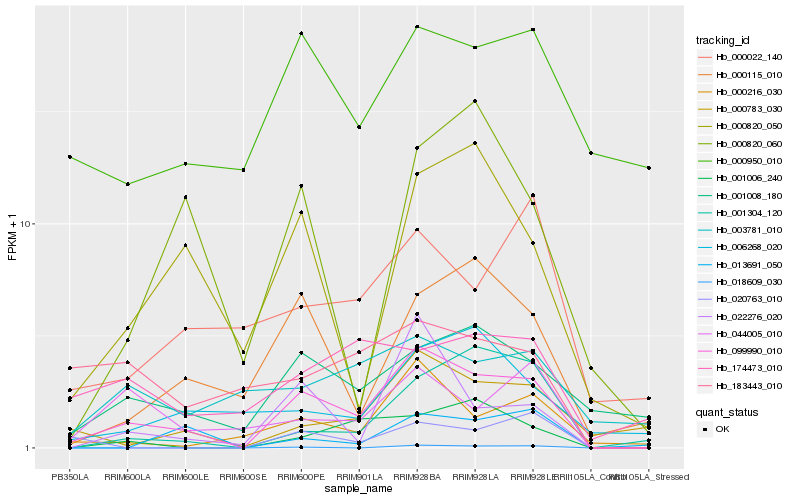
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099990_010 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 11 [Vitis vinifera] |
| 2 |
Hb_000216_030 |
0.2590348273 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 3 |
Hb_001008_180 |
0.2692635973 |
- |
- |
- |
| 4 |
Hb_022276_020 |
0.2693684406 |
- |
- |
- |
| 5 |
Hb_000115_010 |
0.2730324296 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000820_050 |
0.2768542656 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 7 |
Hb_000783_030 |
0.2787301726 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 8 |
Hb_044005_010 |
0.2806161695 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 9 |
Hb_001006_240 |
0.2893143741 |
- |
- |
- |
| 10 |
Hb_001304_120 |
0.2893678584 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 11 |
Hb_006268_020 |
0.30245409 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 12 |
Hb_000022_140 |
0.3033630495 |
- |
- |
hypothetical protein MIMGU_mgv1a009920mg [Erythranthe guttata] |
| 13 |
Hb_000950_010 |
0.3051842312 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 14 |
Hb_018609_030 |
0.30541055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_183443_010 |
0.3085201668 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 16 |
Hb_000820_060 |
0.3087506986 |
- |
- |
- |
| 17 |
Hb_020763_010 |
0.3104040329 |
- |
- |
polyamine oxidase, putative [Ricinus communis] |
| 18 |
Hb_003781_010 |
0.3115302929 |
- |
- |
PREDICTED: histone H2AX [Eucalyptus grandis] |
| 19 |
Hb_174473_010 |
0.3117697536 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 20 |
Hb_013691_050 |
0.3141574168 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |