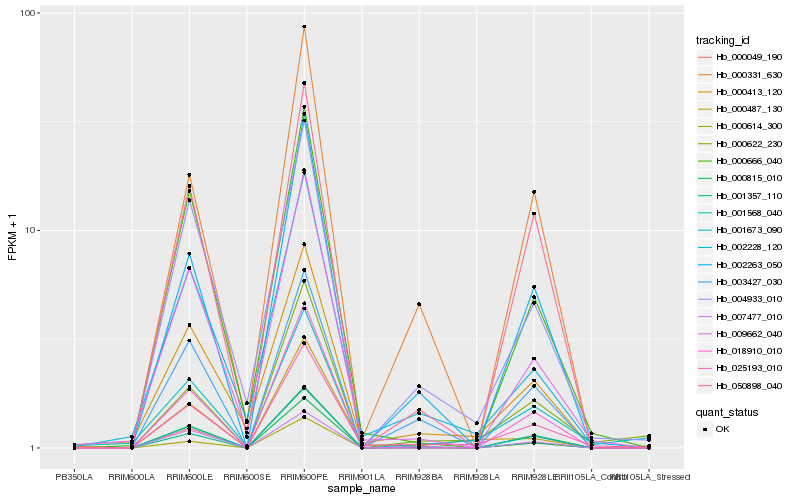
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025193_010 |
0.0 |
- |
- |
PREDICTED: indole-3-acetic acid-amido synthetase GH3.17-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001673_090 |
0.1238500457 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 3 |
Hb_002263_050 |
0.1341023235 |
- |
- |
hypothetical protein POPTR_0016s06160g [Populus trichocarpa] |
| 4 |
Hb_009662_040 |
0.1360986981 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 5 |
Hb_004933_010 |
0.1375239725 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 6 |
Hb_000487_130 |
0.1376205317 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000622_230 |
0.1390965068 |
- |
- |
PREDICTED: uncharacterized protein LOC105642895 [Jatropha curcas] |
| 8 |
Hb_000666_040 |
0.1410640938 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 9 |
Hb_000614_300 |
0.1418772023 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |
| 10 |
Hb_002228_120 |
0.1421118909 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 11 |
Hb_000413_120 |
0.1440490002 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 12 |
Hb_007477_010 |
0.144120799 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 13 |
Hb_001357_110 |
0.144479139 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 14 |
Hb_000331_630 |
0.1456222687 |
- |
- |
hypothetical protein B456_008G203900 [Gossypium raimondii] |
| 15 |
Hb_050898_040 |
0.1460534346 |
- |
- |
- |
| 16 |
Hb_001568_040 |
0.1462694299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000049_190 |
0.148254222 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 18 |
Hb_000815_010 |
0.1490218673 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105117338 [Populus euphratica] |
| 19 |
Hb_018910_010 |
0.1494828345 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 20 |
Hb_003427_030 |
0.1497660381 |
- |
- |
- |