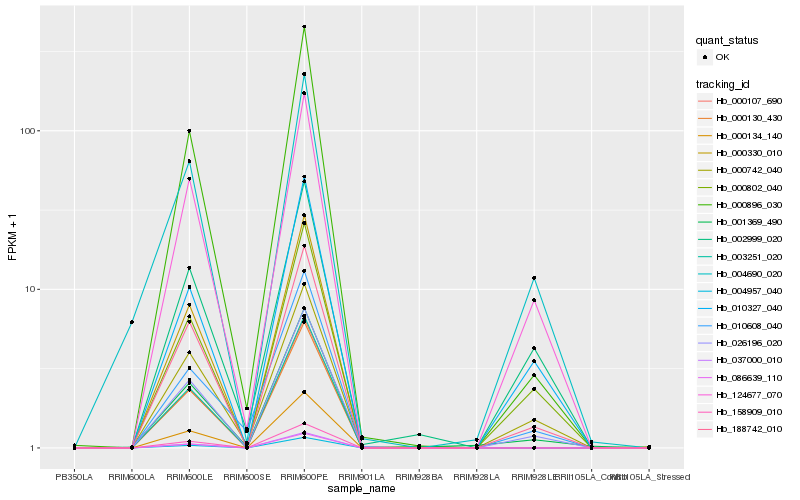
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_490 |
0.0 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 33 [Jatropha curcas] |
| 2 |
Hb_188742_010 |
0.0883868957 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 3 |
Hb_026196_020 |
0.0907652379 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 4 |
Hb_000896_030 |
0.0925690931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_124677_070 |
0.0943891653 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 6 |
Hb_000742_040 |
0.1013133564 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 7 |
Hb_000802_040 |
0.1070876182 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 8 |
Hb_010327_040 |
0.1113485408 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 9 |
Hb_010608_040 |
0.1143034349 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 10 |
Hb_002999_020 |
0.1161571366 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 11 |
Hb_004690_020 |
0.117297455 |
- |
- |
PREDICTED: MLP-like protein 328 [Jatropha curcas] |
| 12 |
Hb_158909_010 |
0.1185550508 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_003251_020 |
0.1185623734 |
- |
- |
Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B [Gossypium arboreum] |
| 14 |
Hb_000107_690 |
0.1185656549 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_037000_010 |
0.1185708462 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 16 |
Hb_004957_040 |
0.1186175386 |
- |
- |
PREDICTED: uncharacterized protein LOC104890338 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_086639_110 |
0.1187045026 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Theobroma cacao] |
| 18 |
Hb_000330_010 |
0.1187122993 |
- |
- |
hypothetical protein POPTR_0005s25020g [Populus trichocarpa] |
| 19 |
Hb_000134_140 |
0.1187733683 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000130_430 |
0.1190693631 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |