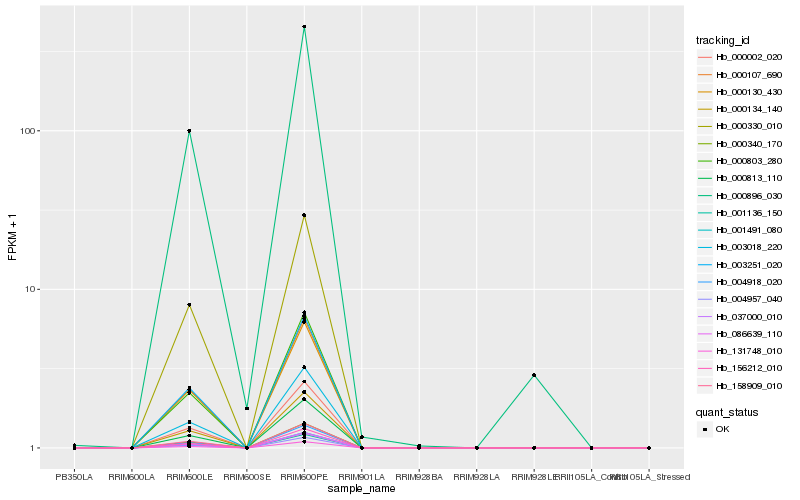
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000896_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_131748_010 |
0.0511444973 |
- |
- |
hypothetical protein PRUPE_ppa022115mg [Prunus persica] |
| 3 |
Hb_000134_140 |
0.051535458 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 4 |
Hb_000340_170 |
0.0516603656 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790-like [Jatropha curcas] |
| 5 |
Hb_156212_010 |
0.0516727083 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_000002_020 |
0.0517886509 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 7 |
Hb_004957_040 |
0.0520628204 |
- |
- |
PREDICTED: uncharacterized protein LOC104890338 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_004918_020 |
0.0520810024 |
- |
- |
- |
| 9 |
Hb_003018_220 |
0.0521436598 |
- |
- |
Snakin-2 [Gossypium arboreum] |
| 10 |
Hb_037000_010 |
0.0524427187 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 11 |
Hb_001491_080 |
0.0529726701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_158909_010 |
0.0531898706 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_000803_280 |
0.0532009862 |
- |
- |
PREDICTED: uncharacterized protein LOC105648326 [Jatropha curcas] |
| 14 |
Hb_003251_020 |
0.0533934067 |
- |
- |
Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B [Gossypium arboreum] |
| 15 |
Hb_000107_690 |
0.0534609108 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_086639_110 |
0.0548519529 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Theobroma cacao] |
| 17 |
Hb_000330_010 |
0.0549075501 |
- |
- |
hypothetical protein POPTR_0005s25020g [Populus trichocarpa] |
| 18 |
Hb_000813_110 |
0.0550594287 |
- |
- |
PREDICTED: uncharacterized protein LOC104884346 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_000130_430 |
0.0569006745 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 20 |
Hb_001136_150 |
0.0572215632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |