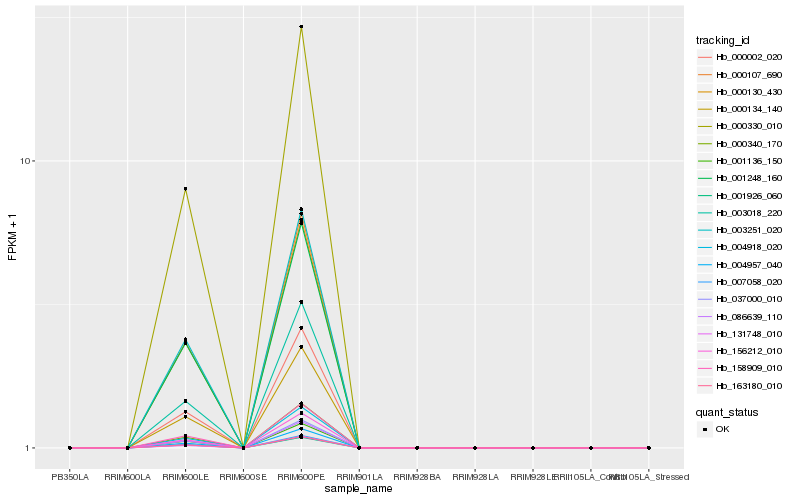
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_037000_010 |
0.0 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 2 |
Hb_004957_040 |
0.0018408267 |
- |
- |
PREDICTED: uncharacterized protein LOC104890338 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_158909_010 |
0.0029913165 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 4 |
Hb_003251_020 |
0.0037124007 |
- |
- |
Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B [Gossypium arboreum] |
| 5 |
Hb_000107_690 |
0.0039448191 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000134_140 |
0.0051695164 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_086639_110 |
0.0081893745 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Theobroma cacao] |
| 8 |
Hb_000330_010 |
0.0083423958 |
- |
- |
hypothetical protein POPTR_0005s25020g [Populus trichocarpa] |
| 9 |
Hb_000130_430 |
0.0132979963 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 10 |
Hb_131748_010 |
0.0134186287 |
- |
- |
hypothetical protein PRUPE_ppa022115mg [Prunus persica] |
| 11 |
Hb_001136_150 |
0.0140213444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001248_160 |
0.0176463057 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 13 |
Hb_000340_170 |
0.0192162496 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790-like [Jatropha curcas] |
| 14 |
Hb_156212_010 |
0.0193011743 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 15 |
Hb_000002_020 |
0.0200576162 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 16 |
Hb_163180_010 |
0.0215684677 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 17 |
Hb_004918_020 |
0.0217195412 |
- |
- |
- |
| 18 |
Hb_003018_220 |
0.0220419891 |
- |
- |
Snakin-2 [Gossypium arboreum] |
| 19 |
Hb_001926_060 |
0.0226718022 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007058_020 |
0.0236505994 |
- |
- |
- |