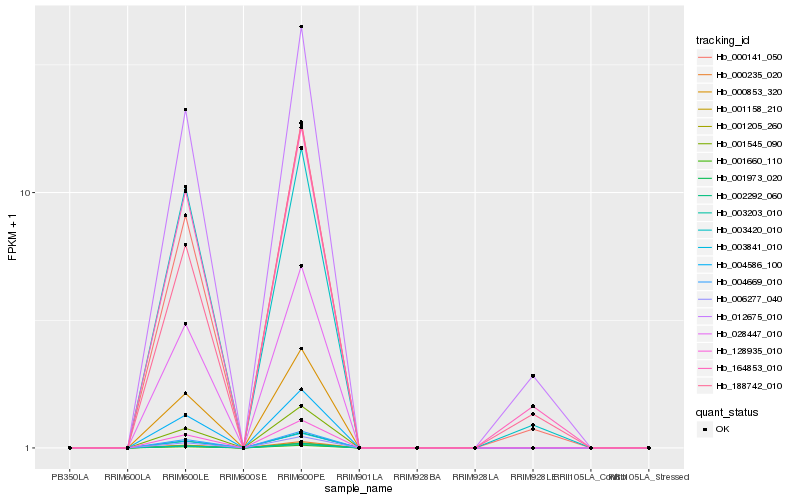
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012675_010 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_164853_010 |
0.0316310669 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 3 |
Hb_000141_050 |
0.038049043 |
- |
- |
hypothetical protein POPTR_0001s00460g [Populus trichocarpa] |
| 4 |
Hb_003420_010 |
0.0812057424 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_188742_010 |
0.0838575163 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 6 |
Hb_001158_210 |
0.0844601698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_128935_010 |
0.0848267899 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Glycine soja] |
| 8 |
Hb_003841_010 |
0.0848543784 |
- |
- |
- |
| 9 |
Hb_004669_010 |
0.0849301905 |
- |
- |
hypothetical protein JCGZ_26481 [Jatropha curcas] |
| 10 |
Hb_002292_060 |
0.0849588874 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 11 |
Hb_000853_320 |
0.0850288214 |
- |
- |
- |
| 12 |
Hb_001660_110 |
0.0851567645 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_006277_040 |
0.0851856015 |
- |
- |
PREDICTED: sodium-dependent phosphate transporter 1-like [Jatropha curcas] |
| 14 |
Hb_001973_020 |
0.0852597976 |
- |
- |
- |
| 15 |
Hb_001205_260 |
0.0853479023 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 16 |
Hb_028447_010 |
0.0855865731 |
- |
- |
hypothetical protein RCOM_1967580 [Ricinus communis] |
| 17 |
Hb_004586_100 |
0.0856665183 |
- |
- |
PREDICTED: uncharacterized protein LOC105120428 isoform X2 [Populus euphratica] |
| 18 |
Hb_000235_020 |
0.0856726982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001545_090 |
0.0864230178 |
- |
- |
PREDICTED: uncharacterized protein LOC103874823 [Brassica rapa] |
| 20 |
Hb_003203_010 |
0.0865375386 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |