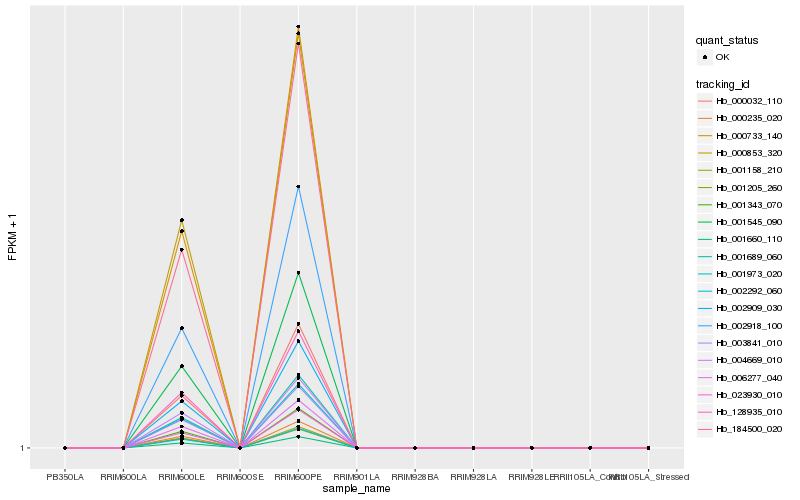
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006277_040 |
0.0 |
- |
- |
PREDICTED: sodium-dependent phosphate transporter 1-like [Jatropha curcas] |
| 2 |
Hb_001660_110 |
0.0002237152 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_001973_020 |
0.0005563344 |
- |
- |
- |
| 4 |
Hb_000853_320 |
0.0012760873 |
- |
- |
- |
| 5 |
Hb_002292_060 |
0.0019009306 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 6 |
Hb_003841_010 |
0.0029213356 |
- |
- |
- |
| 7 |
Hb_128935_010 |
0.003212272 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Glycine soja] |
| 8 |
Hb_000235_020 |
0.0032729129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001545_090 |
0.0072329133 |
- |
- |
PREDICTED: uncharacterized protein LOC103874823 [Brassica rapa] |
| 10 |
Hb_001158_210 |
0.0102679184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002909_030 |
0.0118892369 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1270600 [Ricinus communis] |
| 12 |
Hb_001343_070 |
0.0120712445 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62680, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000733_140 |
0.0155840844 |
- |
- |
PREDICTED: ras-related protein Rab2BV-like [Jatropha curcas] |
| 14 |
Hb_001689_060 |
0.0179561544 |
- |
- |
PREDICTED: uncharacterized protein LOC103846154 [Brassica rapa] |
| 15 |
Hb_004669_010 |
0.0201690577 |
- |
- |
hypothetical protein JCGZ_26481 [Jatropha curcas] |
| 16 |
Hb_002918_100 |
0.0219491778 |
- |
- |
hypothetical protein JCGZ_21466 [Jatropha curcas] |
| 17 |
Hb_000032_110 |
0.0220681582 |
- |
- |
nadhp hc toxin reductase, putative [Ricinus communis] |
| 18 |
Hb_001205_260 |
0.0235232531 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 19 |
Hb_023930_010 |
0.0239167176 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 20 |
Hb_184500_020 |
0.0249029431 |
- |
- |
- |