| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
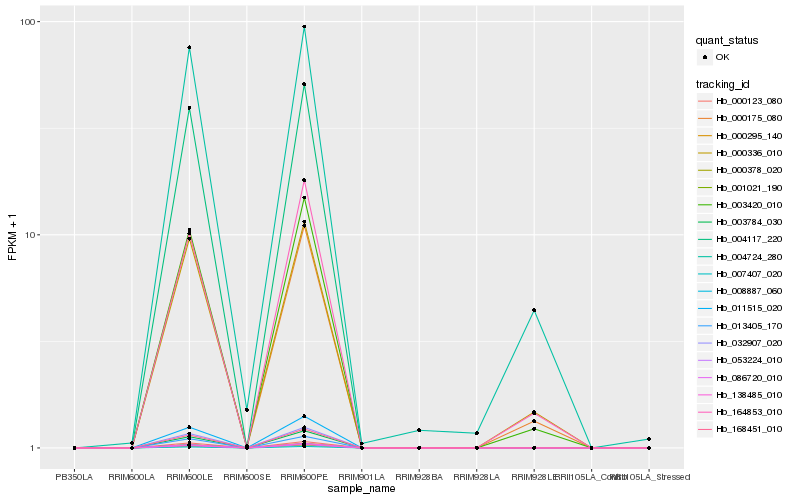
Hb_003420_010 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_164853_010 |
0.0563364234 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 3 |
Hb_000336_010 |
0.0648537034 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_000175_080 |
0.0673890714 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 5 |
Hb_086720_010 |
0.0697222237 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 6 |
Hb_007407_020 |
0.0697387984 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_000295_140 |
0.0697477148 |
- |
- |
PREDICTED: uncharacterized protein LOC105628741 [Jatropha curcas] |
| 8 |
Hb_032907_020 |
0.0697759588 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_053224_010 |
0.0701786944 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 10 |
Hb_168451_010 |
0.0702163036 |
- |
- |
PREDICTED: acid phosphatase 1-like [Populus euphratica] |
| 11 |
Hb_138485_010 |
0.0703522347 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 12 |
Hb_003784_030 |
0.0715981421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008887_060 |
0.0722170544 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001021_190 |
0.0722930275 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 15 |
Hb_011515_020 |
0.0730427457 |
- |
- |
PREDICTED: dehydration-responsive element-binding protein 1F-like [Jatropha curcas] |
| 16 |
Hb_004117_220 |
0.0739847663 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 17 |
Hb_000378_020 |
0.0740660638 |
- |
- |
PREDICTED: uncharacterized protein LOC101497268 [Cicer arietinum] |
| 18 |
Hb_004724_280 |
0.0743342976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000123_080 |
0.0744330945 |
transcription factor |
TF Family: B3 |
hypothetical protein VITISV_040593 [Vitis vinifera] |
| 20 |
Hb_013405_170 |
0.0746624126 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |