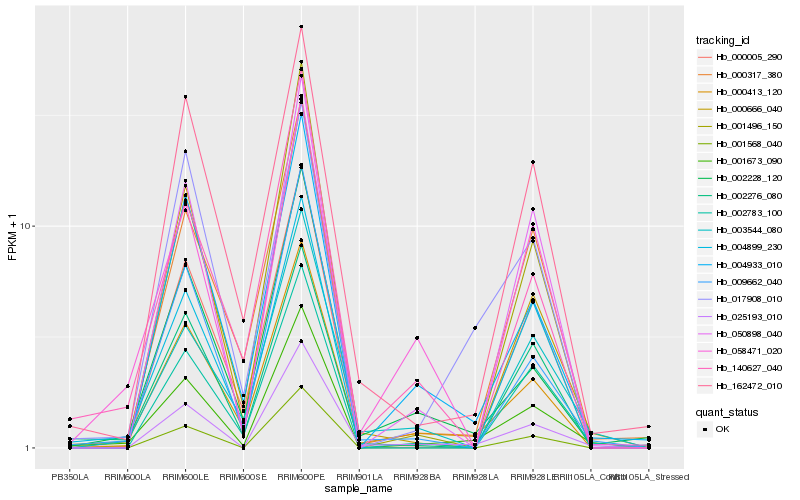
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001673_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 2 |
Hb_000413_120 |
0.1222888657 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 3 |
Hb_025193_010 |
0.1238500457 |
- |
- |
PREDICTED: indole-3-acetic acid-amido synthetase GH3.17-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_009662_040 |
0.1384895403 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 5 |
Hb_004933_010 |
0.1410679661 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 6 |
Hb_002783_100 |
0.1455809275 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 7 |
Hb_002228_120 |
0.1489906161 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 8 |
Hb_000317_380 |
0.1516473169 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 9 |
Hb_050898_040 |
0.1539987964 |
- |
- |
- |
| 10 |
Hb_000666_040 |
0.1544103031 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 11 |
Hb_140627_040 |
0.1555850171 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 12 |
Hb_001496_150 |
0.1572146244 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 13 |
Hb_004899_230 |
0.1577412503 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 14 |
Hb_000005_290 |
0.1580205698 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_003544_080 |
0.1585476809 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 16 |
Hb_002276_080 |
0.1590433317 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 17 |
Hb_058471_020 |
0.1599043615 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 18 |
Hb_017908_010 |
0.1599998418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_162472_010 |
0.1627862643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001568_040 |
0.1635368104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |