| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
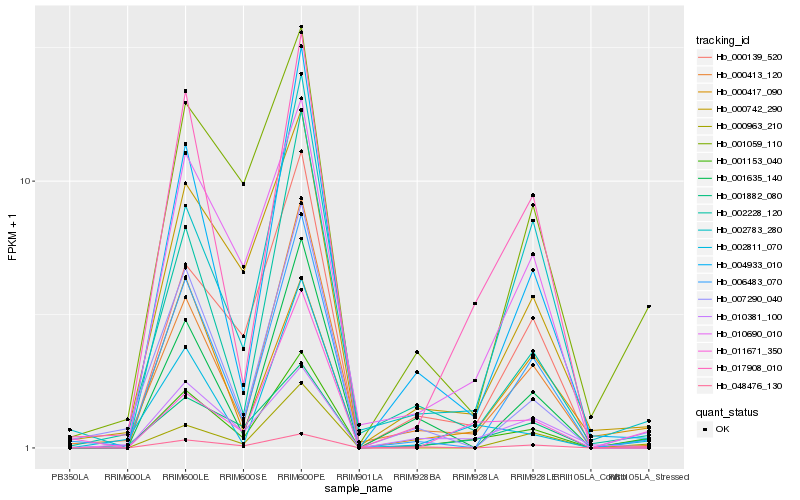
Hb_001153_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105128973 [Populus euphratica] |
| 2 |
Hb_048476_130 |
0.1721569593 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 3 |
Hb_017908_010 |
0.1746291611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004933_010 |
0.1769425738 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 5 |
Hb_000413_120 |
0.1785110596 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 6 |
Hb_006483_070 |
0.1796499365 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_000742_290 |
0.1797702308 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 8 |
Hb_001059_110 |
0.1816285208 |
- |
- |
PREDICTED: uncharacterized protein LOC105649457 [Jatropha curcas] |
| 9 |
Hb_000963_210 |
0.1824035426 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 10 |
Hb_002811_070 |
0.1855747766 |
- |
- |
Cytidine/deoxycytidylate deaminase family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_010690_010 |
0.1877806733 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 12 |
Hb_001882_080 |
0.1893772985 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103324347 [Prunus mume] |
| 13 |
Hb_011671_350 |
0.1894954282 |
- |
- |
PREDICTED: uncharacterized protein LOC105648659 [Jatropha curcas] |
| 14 |
Hb_001635_140 |
0.1904687374 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_002783_280 |
0.1916235703 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 16 |
Hb_000417_090 |
0.1917966981 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_010381_100 |
0.1924847621 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 18 |
Hb_000139_520 |
0.1930225148 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 19 |
Hb_002228_120 |
0.1947983332 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 20 |
Hb_007290_040 |
0.195047682 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |