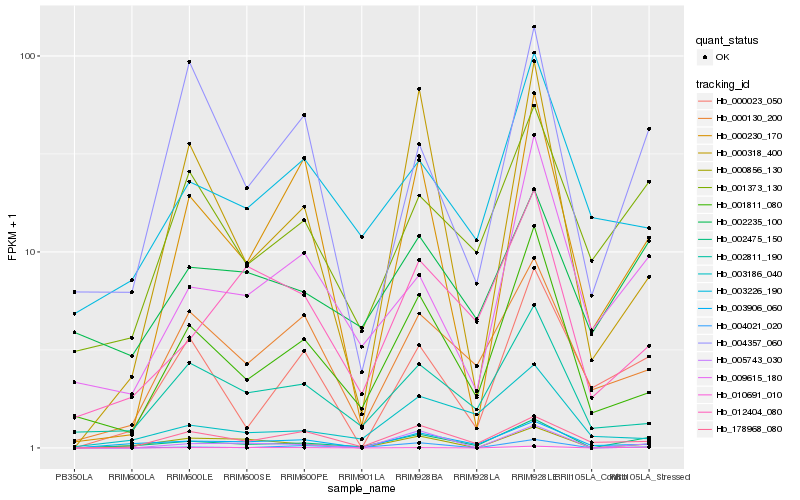
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002475_150 |
0.0 |
- |
- |
PREDICTED: importin subunit beta-3-like [Jatropha curcas] |
| 2 |
Hb_001811_080 |
0.2247843839 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 3 |
Hb_001373_130 |
0.2271189537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003906_060 |
0.2277535813 |
- |
- |
hypothetical protein VITISV_022975 [Vitis vinifera] |
| 5 |
Hb_009615_180 |
0.2345570134 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 6 |
Hb_005743_030 |
0.2437787224 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 7 |
Hb_000856_130 |
0.2473890499 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_010691_010 |
0.24924616 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 9 |
Hb_000023_050 |
0.2505820246 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 10 |
Hb_012404_080 |
0.2512963289 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 11 |
Hb_002235_100 |
0.2537604897 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002811_190 |
0.2554198851 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 13 |
Hb_000130_200 |
0.2557167529 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 14 |
Hb_000318_400 |
0.2573442188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_178968_080 |
0.259455031 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000230_170 |
0.2595660171 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 17 |
Hb_004021_020 |
0.2598390413 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 18 |
Hb_004357_060 |
0.2608251203 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 19 |
Hb_003226_190 |
0.2635867226 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 20 |
Hb_003186_040 |
0.2643806229 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |