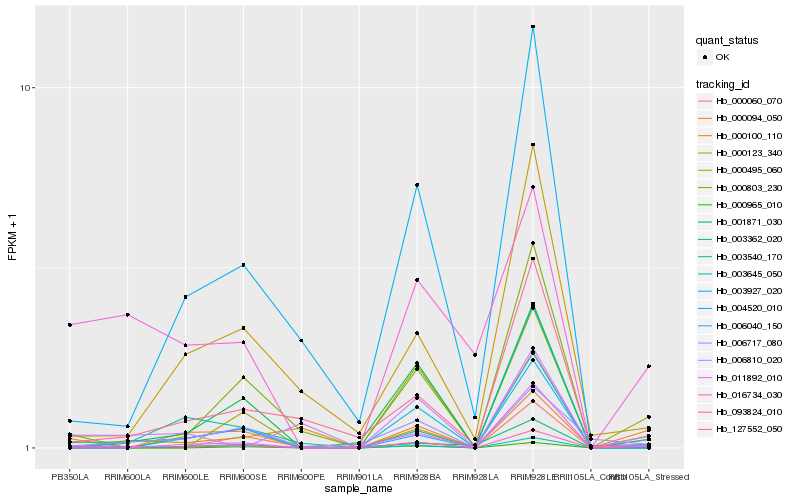
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093824_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103326132 [Prunus mume] |
| 2 |
Hb_000094_050 |
0.2137483416 |
- |
- |
- |
| 3 |
Hb_016734_030 |
0.2661464243 |
- |
- |
- |
| 4 |
Hb_003645_050 |
0.2682491834 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000495_060 |
0.2713196629 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 6 |
Hb_001871_030 |
0.2735265261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006717_080 |
0.2747854208 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 8 |
Hb_011892_010 |
0.2922856732 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000060_070 |
0.2930428947 |
- |
- |
hypothetical protein EUTSA_v10023188mg, partial [Eutrema salsugineum] |
| 10 |
Hb_000100_110 |
0.2979739175 |
- |
- |
- |
| 11 |
Hb_003362_020 |
0.2993397657 |
- |
- |
PREDICTED: uncharacterized protein LOC104098435, partial [Nicotiana tomentosiformis] |
| 12 |
Hb_003540_170 |
0.3036025014 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21705, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000123_340 |
0.3037348254 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 14 |
Hb_000965_010 |
0.3040571864 |
- |
- |
hypothetical protein VITISV_016917 [Vitis vinifera] |
| 15 |
Hb_006040_150 |
0.3067564784 |
- |
- |
- |
| 16 |
Hb_000803_230 |
0.3069676846 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 17 |
Hb_127552_050 |
0.3083363625 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 18 |
Hb_006810_020 |
0.3096591861 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Jatropha curcas] |
| 19 |
Hb_004520_010 |
0.313042796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 20 |
Hb_003927_020 |
0.3134075516 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |